Fisher SSF
John Fieberg
Preamble
Load libraries
library(ezknitr)
library(knitr)
library(lubridate)
library(raster)
library(move)
library(amt)
library(tidyverse)
options(width=150)
opts_chunk$set(fig.width=12,fig.height=4.5)Read in annotated available data for RSF modeling
ssfdat<-read.csv("data/AllStepsFisher2018-EnvDATA-results.csv")Convert time variables
ssfdat$t1_<-as.POSIXct(ssfdat$t1_, format="%Y-%m-%d %H:%M:%OS", tz="UTC")
ssfdat$t2_<-as.POSIXct(ssfdat$t2_, format="%Y-%m-%d %H:%M:%OS", tz="UTC")Simplify some variable names and make case a numeric variable
names(ssfdat)[c(16,21,20,22)]<-c("id","Elevation", "LandClass", "PopDens")
ssfdat$case_<-as.numeric(ssfdat$case)Create landcover classes (as suggested by Scott Lapoint :)
ssfdat$LandClass<-as.character(ssfdat$LandClass)
ssfdat<-ssfdat %>% mutate(landC = fct_collapse(LandClass,
agri = c("11", "14", "30"),
forest =c("30","40","50","60", "70","80", "90","100"),
shrub= c("110", "130", "150"),
grass = c("120", "140"),
wet= c("160"),
other = c("170", "180", "190", "200", "210", "220")))## Warning: Unknown levels in `f`: 11, 14, 40, 60, 80, 90, 150, 170, 180, 200, 220Center and scale variables
ssfdat<-ssfdat %>% mutate(elev=as.numeric(scale(Elevation)),
popD=as.numeric(scale(PopDens)))Explore the data:
Look Distribution of variables for used and available
binomial_smooth <- function(...) {
geom_smooth(method = "glm", method.args = list(family = "binomial"), ...)
}
ggplot(ssfdat,aes(x=Elevation, y=case_))+
stat_smooth(method="glm", method.args = list(family = "binomial"))+
binomial_smooth(formula = y ~ splines::ns(x, 5), colour="red")+
facet_wrap(~id, scales="free")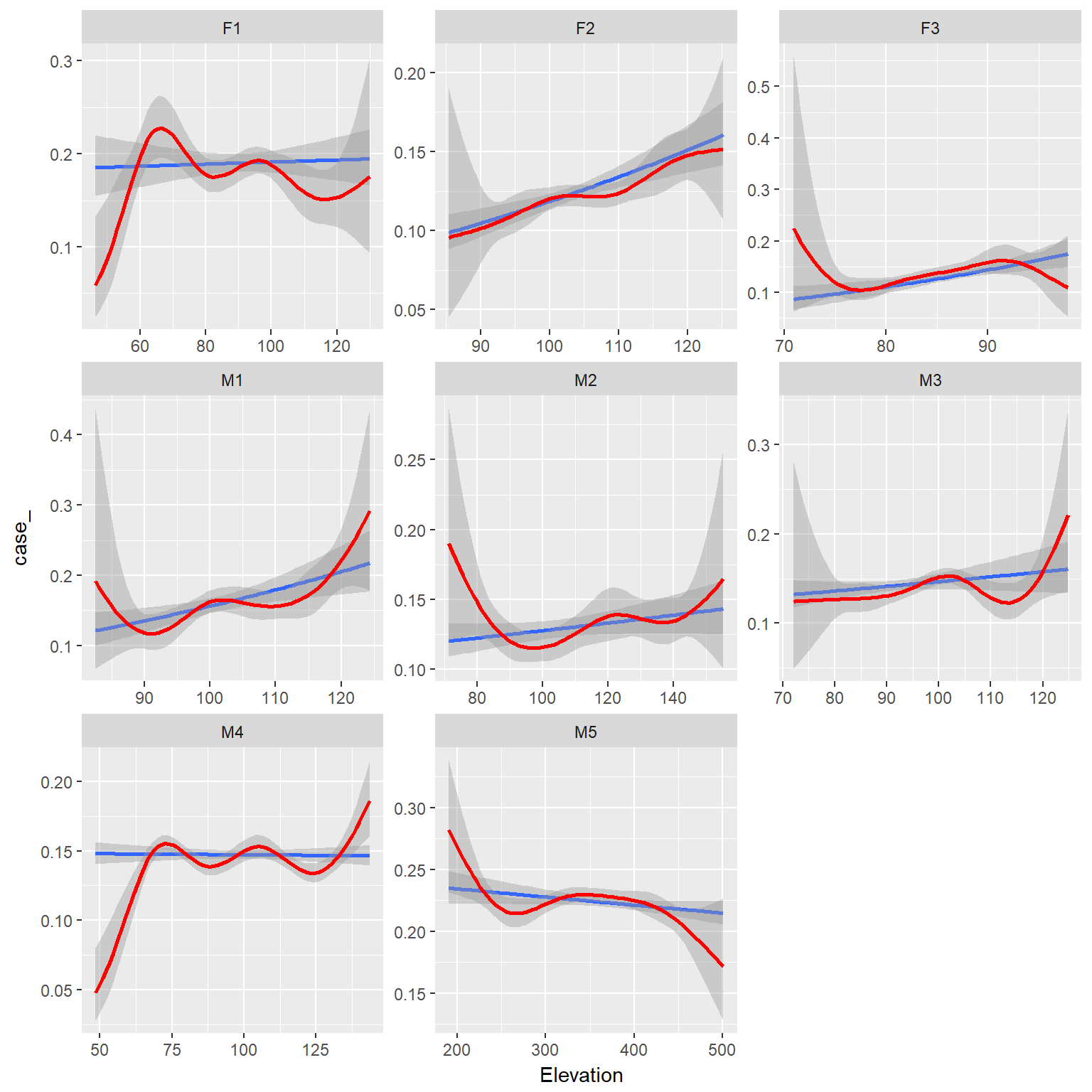
ggplot(ssfdat,aes(x=PopDens, y=case_))+
stat_smooth(method="glm", method.args = list(family = "binomial"))+
binomial_smooth(formula = y ~ splines::ns(x, 5), colour="red")+
facet_wrap(~id, scales="free")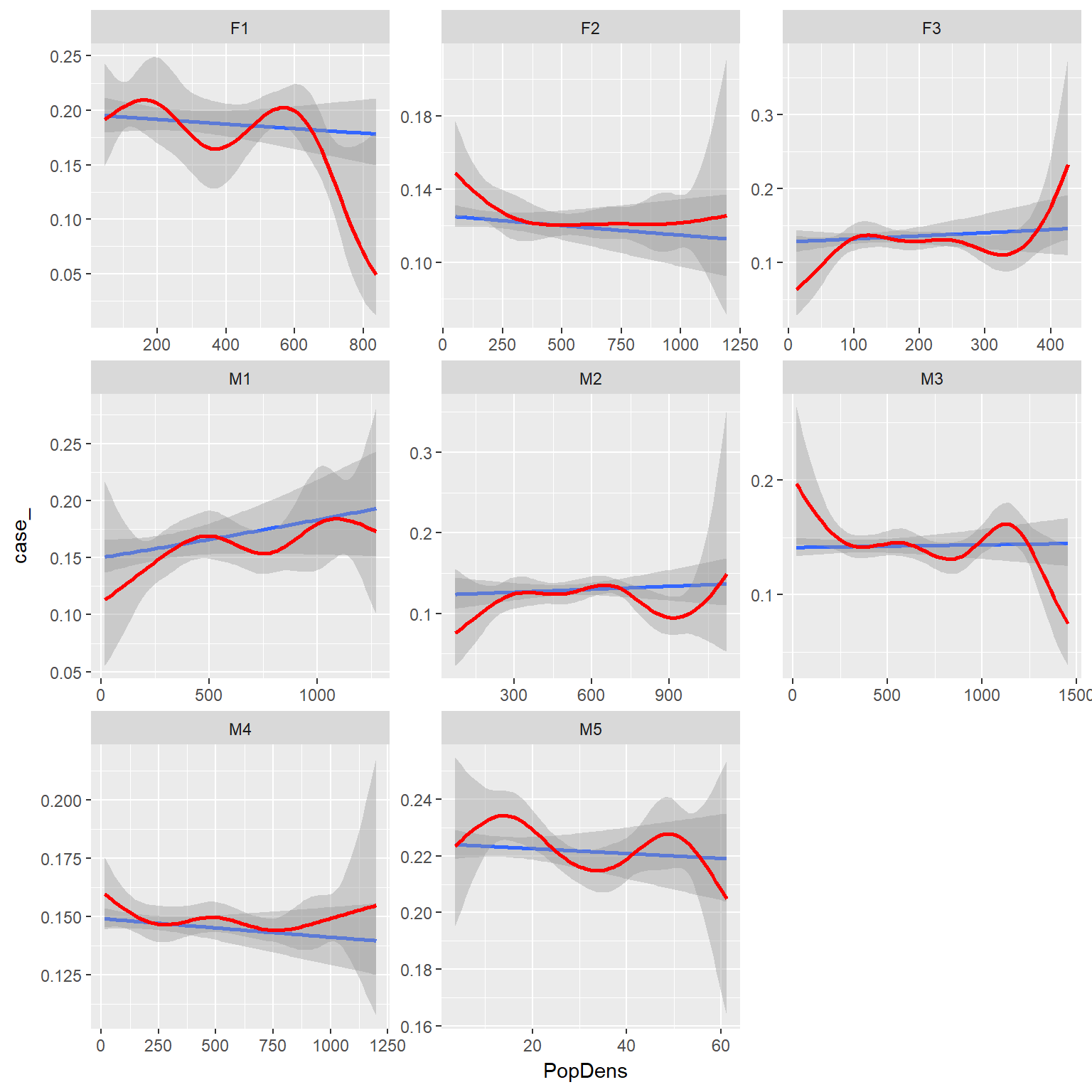
ggplot(ssfdat, aes(x=landC, y=..prop..,group=case_, colour=case_))+
geom_bar(position="dodge", aes(fill=case_))+facet_wrap(~id, scales="free")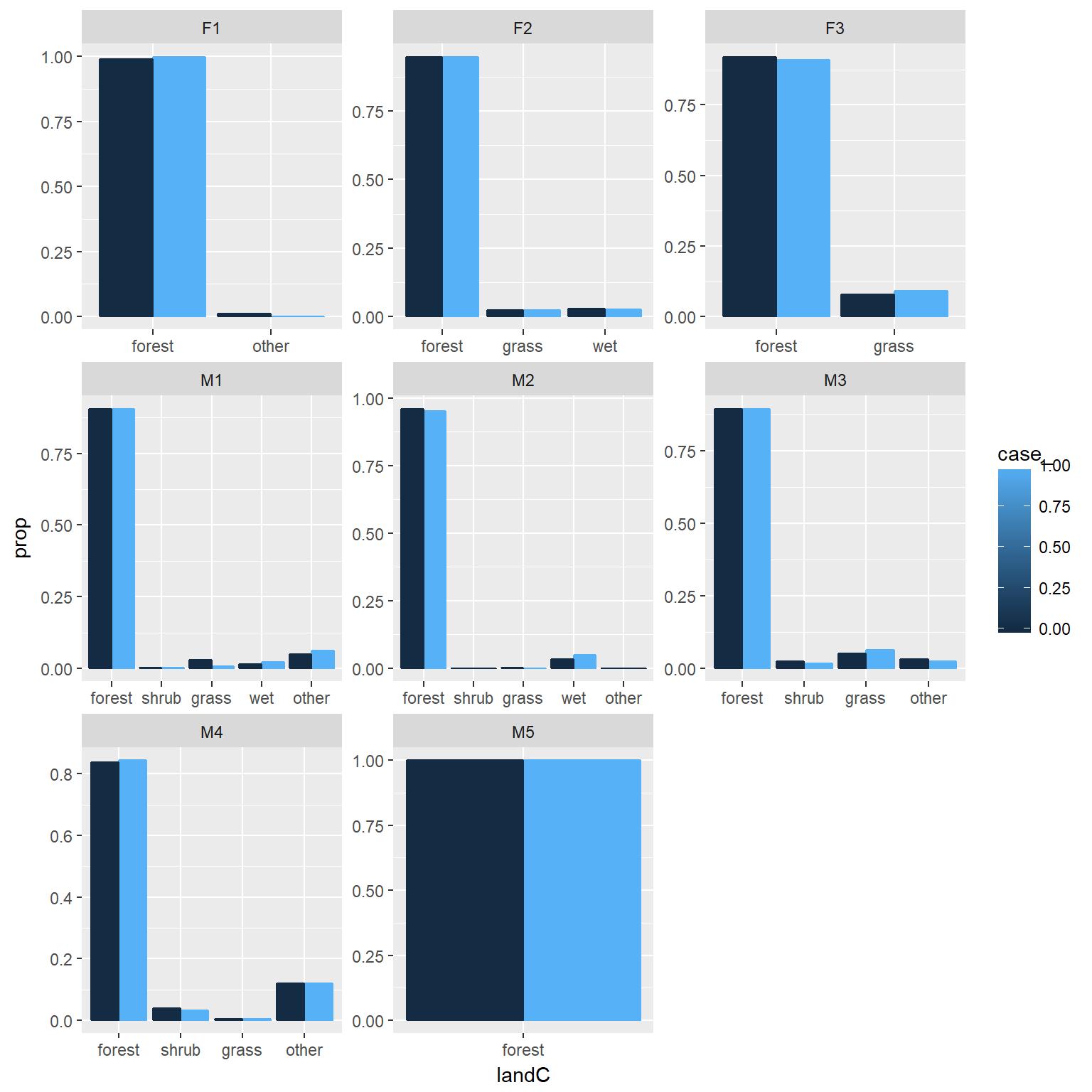
Now, fit an RSF model to data from each animal. Since not all animals experience all habitat types, lets just explore forest versus non-forest
ssfdat$forest<-ifelse(ssfdat$landC=="forest", 1, 0)Fit an SSF to a single animal
summary(fit_issf(case_ ~ Elevation+PopDens+forest+sl_+log(sl_)+strata(step_id_),
data = subset(ssfdat, id=="M1")))## Call:
## coxph(formula = Surv(rep(1, 4697L), case_) ~ Elevation + PopDens +
## forest + sl_ + log(sl_) + strata(step_id_), data = data,
## method = "exact")
##
## n= 4697, number of events= 748
##
## coef exp(coef) se(coef) z Pr(>|z|)
## Elevation 0.0775860 1.0806752 0.0151523 5.120 3.05e-07 ***
## PopDens 0.0005830 1.0005832 0.0008624 0.676 0.499
## forest -0.2186602 0.8035947 0.2767507 -0.790 0.429
## sl_ 0.0035169 1.0035230 0.0005275 6.666 2.62e-11 ***
## log(sl_) 0.5099396 1.6651906 0.0431510 11.818 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## Elevation 1.0807 0.9253 1.0491 1.113
## PopDens 1.0006 0.9994 0.9989 1.002
## forest 0.8036 1.2444 0.4672 1.382
## sl_ 1.0035 0.9965 1.0025 1.005
## log(sl_) 1.6652 0.6005 1.5301 1.812
##
## Rsquare= 0.161 (max possible= 0.402 )
## Likelihood ratio test= 823 on 5 df, p=0
## Wald test = 474.4 on 5 df, p=0
## Score (logrank) test = 924.8 on 5 df, p=0Note, animal 1587 was always in forest, so we can’t include forest for this individual. Fit an SSF model to data from each animal
fitted_ssf <- function(data){
n0<-sum(data$forest==0)
if(n0==0){ # this will be id = M5
mod <- fit_issf(case_ ~ Elevation+PopDens+sl_+log(sl_)+strata(step_id_), data=data)
}
if(n0>0){
mod <- fit_issf(case_ ~ Elevation+PopDens+forest+sl_+log(sl_)+strata(step_id_), data=data)
}
return(mod)
}
ssffits <-ssfdat %>% nest(-id) %>%
dplyr::mutate(mod = purrr::map(data, fitted_ssf)) ## Warning in fitter(X, Y, strats, offset, init, control, weights = weights, : Loglik converged before variable 3 ; beta may be infinite.Look at first model
ssffits$mod[[1]]## $model
## Call:
## survival::clogit(formula, data = data, ...)
##
## coef exp(coef) se(coef) z p
## Elevation 0.003495 1.003501 0.002111 1.66 0.098
## PopDens -0.023165 0.977101 0.011013 -2.10 0.035
## sl_ 0.004443 1.004453 0.000712 6.24 4.3e-10
## log(sl_) 0.848218 2.335482 0.026823 31.62 < 2e-16
##
## Likelihood ratio test=6522 on 4 df, p=0
## n= 52303, number of events= 11670
##
## $sl_
## NULL
##
## $ta_
## NULL
##
## $more
## NULL
##
## attr(,"class")
## [1] "fit_clogit" "list"Now, use tidy to extract information about the model fits
ssffits <- ssffits %>%
dplyr::mutate(tidy = purrr::map(mod, ~broom::tidy(.x$model)),
n = purrr::map(data, nrow) %>% simplify())
ssffits$tidy## [[1]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.003495026 0.0021107020 1.655859 9.775030e-02 -0.0006418742 0.007631925
## 2 PopDens -0.023165483 0.0110130481 -2.103458 3.542576e-02 -0.0447506609 -0.001580306
## 3 sl_ 0.004442938 0.0007115682 6.243869 4.268785e-10 0.0030482903 0.005837586
## 4 log(sl_) 0.848218389 0.0268226202 31.623249 0.000000e+00 0.7956470195 0.900789759
##
## [[2]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.048538780 0.0111738942 4.343945 1.399467e-05 0.026638349 7.043921e-02
## 2 PopDens -0.001573539 0.0007695850 -2.044659 4.088847e-02 -0.003081898 -6.518015e-05
## 3 forest 0.469534741 0.1541960249 3.045051 2.326410e-03 0.167316085 7.717534e-01
## 4 sl_ 0.002716978 0.0007430072 3.656732 2.554515e-04 0.001260711 4.173245e-03
## 5 log(sl_) 0.532056011 0.0398435941 13.353615 0.000000e+00 0.453964001 6.101480e-01
##
## [[3]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.0868248699 0.017981094 4.8286756 1.374441e-06 0.051582573 0.122067167
## 2 PopDens -0.0022533285 0.002703087 -0.8336130 4.044991e-01 -0.007551281 0.003044624
## 3 forest -0.5905269701 0.307511601 -1.9203405 5.481491e-02 -1.193238632 0.012184692
## 4 sl_ 0.0007653481 0.001431207 0.5347571 5.928178e-01 -0.002039766 0.003570462
## 5 log(sl_) 0.6034485710 0.060744413 9.9342234 0.000000e+00 0.484391709 0.722505433
##
## [[4]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.029150946 0.0071107376 4.0995672 4.139235e-05 0.01521416 0.043087736
## 2 PopDens -0.002583034 0.0009086062 -2.8428536 4.471160e-03 -0.00436387 -0.000802199
## 3 forest -0.258175972 0.3729681440 -0.6922199 4.887992e-01 -0.98918010 0.472828158
## 4 sl_ 0.001929604 0.0004162594 4.6355796 3.559387e-06 0.00111375 0.002745457
## 5 log(sl_) 0.441261838 0.0307360453 14.3564936 0.000000e+00 0.38102030 0.501503380
##
## [[5]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.033738604 9.595590e-03 3.51605325 0.0004380131 0.014931594 0.0525456138
## 2 PopDens -0.002084688 1.477291e-03 -1.41115664 0.1581984331 -0.004980125 0.0008107479
## 3 forest 17.394896350 1.592353e+03 0.01092402 0.9912840642 -Inf Inf
## 4 sl_ 0.002719498 8.442206e-04 3.22131266 0.0012760486 0.001064857 0.0043741404
## 5 log(sl_) 0.751101181 4.694472e-02 15.99969348 0.0000000000 0.659091214 0.8431111472
##
## [[6]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.0775859914 0.0151523355 5.1203982 3.048912e-07 0.047887960 0.107284023
## 2 PopDens 0.0005830073 0.0008623567 0.6760628 4.990008e-01 -0.001107181 0.002273195
## 3 forest -0.2186601922 0.2767506759 -0.7900981 4.294705e-01 -0.761081550 0.323761165
## 4 sl_ 0.0035168540 0.0005275455 6.6664470 2.620704e-11 0.002482884 0.004550824
## 5 log(sl_) 0.5099395721 0.0431510289 11.8175530 0.000000e+00 0.425365110 0.594514035
##
## [[7]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.0152899141 0.0045019257 3.3963053 0.0006830213 0.0064663019 0.0241135264
## 2 PopDens -0.0005107236 0.0006208297 -0.8226469 0.4107088105 -0.0017275275 0.0007060802
## 3 forest 0.2126658070 0.0927658171 2.2925018 0.0218767012 0.0308481464 0.3944834676
## 4 sl_ 0.0019746189 0.0006740995 2.9292693 0.0033975991 0.0006534082 0.0032958296
## 5 log(sl_) 0.6435958428 0.0243878417 26.3900287 0.0000000000 0.5957965514 0.6913951342
##
## [[8]]
## term estimate std.error statistic p.value conf.low conf.high
## 1 Elevation 0.046735632 0.0073259190 6.379491 1.776779e-10 0.03237709 0.0610941694
## 2 PopDens -0.001119928 0.0008470060 -1.322220 1.860949e-01 -0.00278003 0.0005401729
## 3 forest -0.274434455 0.1972930455 -1.390999 1.642257e-01 -0.66112172 0.1122528083
## 4 sl_ -0.003383690 0.0007719734 -4.383169 1.169651e-05 -0.00489673 -0.0018706501
## 5 log(sl_) 0.687138356 0.0411194079 16.710804 0.000000e+00 0.60654580 0.7677309147Now, create data frame w/ the coefficients, etc
ssf_coefs <- ssffits %>%
tidyr::unnest(tidy) %>%
dplyr::select(-(std.error:conf.high))
ssf_coefs %>% tidyr::spread(term, estimate)## # A tibble: 8 x 7
## id n Elevation forest `log(sl_)` PopDens sl_
## <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 F1 6391 0.0337 17.4 0.751 -0.00208 0.00272
## 2 F2 22165 0.0467 -0.274 0.687 -0.00112 -0.00338
## 3 F3 10169 0.0868 -0.591 0.603 -0.00225 0.000765
## 4 M1 4697 0.0776 -0.219 0.510 0.000583 0.00352
## 5 M2 10709 0.0292 -0.258 0.441 -0.00258 0.00193
## 6 M3 15547 0.0485 0.470 0.532 -0.00157 0.00272
## 7 M4 54810 0.0153 0.213 0.644 -0.000511 0.00197
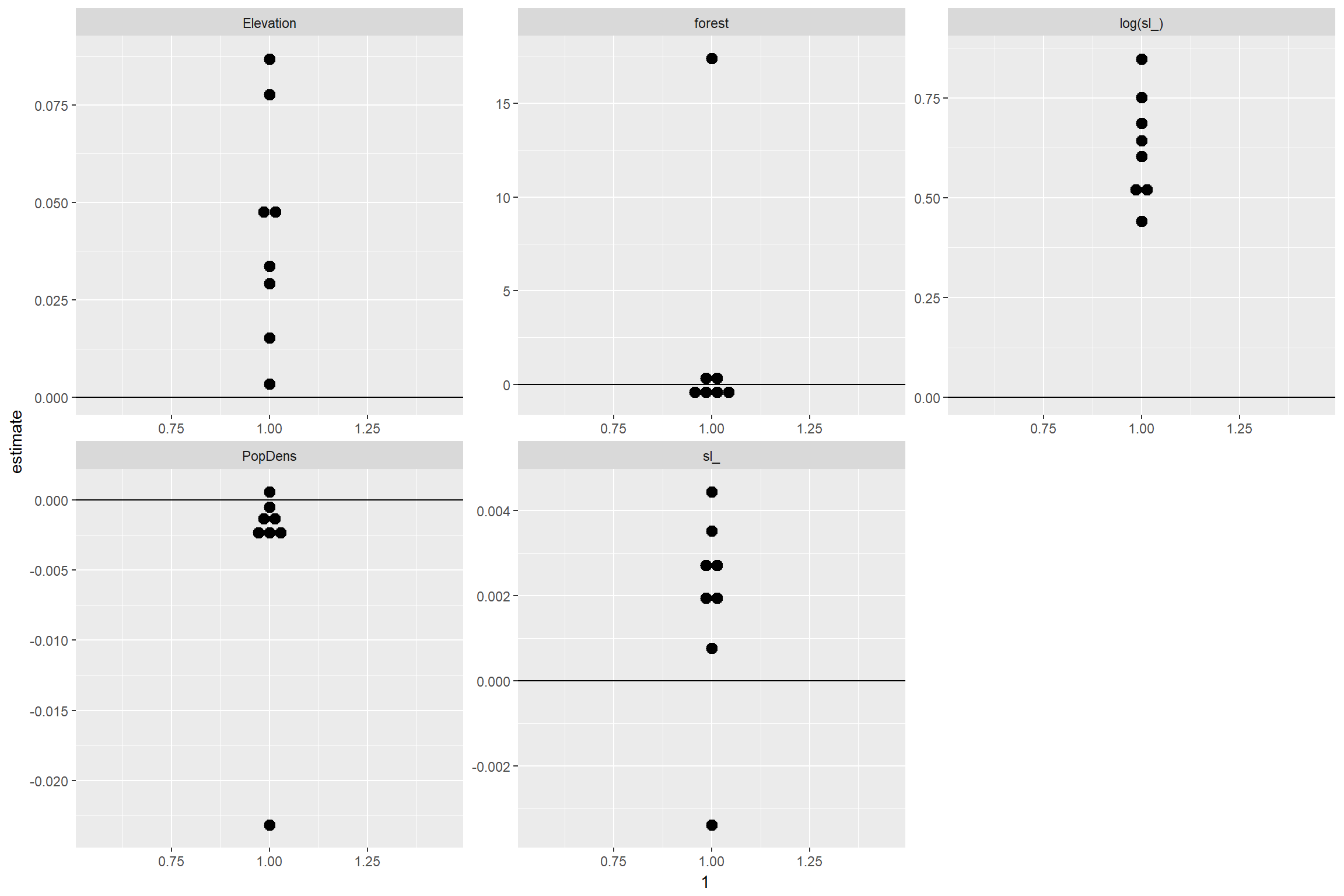
## 8 M5 52303 0.00350 NA 0.848 -0.0232 0.00444Plot coefficients
ssf_coefs %>%
ggplot(., aes(x=1, y=estimate)) +
geom_dotplot(binaxis="y", stackdir="center")+geom_hline(yintercept=0)+
facet_wrap(~term, scales="free")## `stat_bindot()` using `bins = 30`. Pick better value with `binwidth`.