TestVignette
John Fieberg
Purpose
- test whether you have everything set up for the workshop
- retrieve and explore animal movement data from Movebank
- illustrate how to work with functions in the animal movement tools package (amt)
- write out available points for further annotating in EnvDATA
Preamble
Load libraries
library(knitr)
library(lubridate)
library(maptools)
library(raster)
library(move)
library(amt)
library(ggmap)
library(tibble)
library(leaflet)
library(dplyr)
options(width=165,digits.secs = 3)
opts_chunk$set(fig.width=12,fig.height=4.5, error=TRUE,cache = F)Record time for running all code
ptm<-proc.time()Set the seed for the random number generator, so it will be possible to reproduce the random points
set.seed(10299)Create a login object for a user account at movebank.org
loginStored <- movebankLogin(username="MovebankWorkshop", password="genericuserforthisexercise")Get overview information about a Movebank study. Be sure to check the citation and license terms if not using your own data.
getMovebankStudy(study="Martes pennanti LaPoint New York", login=loginStored) # see study-level info## acknowledgements
## 1 This work was supported in part by NSF (grant 0756920 to RK), the New York State Museum, \nthe Max Planck Institute for Ornithology; the North Carolina Museum of Natural Sciences, and a National Geographic Society Waitt Grant (SDL). We thank the Albany Pine Bush Preserve, Dina Dechmann, Neil Gifford, Wolfgang Heidrich, Bart Kranstauber, Franz Kümmeth, Roger Powell, Kamran Safi, Marco Smolla, and Brad Stratton for their logistical support and valuable input.
## bounding_box
## 1 NA
## citation
## 1 LaPoint, S, Gallery P, Wikelski M, Kays R (2013) Animal behavior, cost-based corridor models, and real corridors. Landscape Ecology, v 28 i 8, p 1615–1630. doi:10.1007/s10980-013-9910-0
## comments enable_for_animal_tracker grants_used has_quota i_am_owner
## 1 NA NA National Science Foundation (#0756920 to RWK) and the National Geographic Society's Waitt Grant (to SDL) true false
## id
## 1 6925808
## license_terms
## 1 These data have been published by the Movebank Data Repository with DOI 10.5441/001/1.2tp2j43g. See www.datarepository.movebank.org/handle/10255/move.328. Please contact the PI prior to use of these data for any purposes.
## location_description main_location_lat main_location_long name number_of_deployments number_of_events number_of_individuals
## 1 NA 42.75205 -73.86246 Martes pennanti LaPoint New York NA NA NA
## number_of_tags principal_investigator_address principal_investigator_email principal_investigator_name
## 1 NA NA NA NA
## study_objective
## 1 To investigate the use of corridors by fisher within a suburban landscape and to validate cost-based corridor model predictions with both animal tracking data and camera traps.
## study_type suspend_license_terms timestamp_end timestamp_start i_can_see_data there_are_data_which_i_cannot_see
## 1 research false NA NA true falseYou may receive a message that you do not have access to data in a study. If you are not a data manager for a study, the owner may require that you read and accept the license terms once before you may download it. Currently license terms cannot be accepted directly from the move package. To view and accept them, go to movebank.org, search for the study and begin a download, and view and accept the license terms. After that you will be able to load from R. Load data from a study in Movebank and create a MoveStack object. For more details and options see https://cran.r-project.org/web/packages/move/index.html.
fisher.move <- getMovebankData(study="Martes pennanti LaPoint New York", login=loginStored)
head(fisher.move)## sensor_type_id behavioural_classification eobs_battery_voltage eobs_fix_battery_voltage eobs_horizontal_accuracy_estimate eobs_key_bin_checksum
## 5 653 NA NA NA NA
## 6 653 NA NA NA NA
## 7 653 NA NA NA NA
## 9 653 NA NA NA NA
## 15 653 NA NA NA NA
## 16 653 NA NA NA NA
## eobs_speed_accuracy_estimate eobs_start_timestamp eobs_status eobs_temperature eobs_type_of_fix eobs_used_time_to_get_fix ground_speed heading
## 5 NA NA NA NA NA 80 NA NA
## 6 NA NA NA NA NA 7 NA NA
## 7 NA NA NA NA NA 58 NA NA
## 9 NA NA NA NA NA 90 NA NA
## 15 NA NA NA NA NA 71 NA NA
## 16 NA NA NA NA NA 57 NA NA
## height_above_ellipsoid location_lat location_long manually_marked_outlier timestamp update_ts deployment_id event_id tag_id
## 5 NA 42.79528 -73.86015 NA 2011-02-11 18:06:13.999 1970-01-01 00:00:00.000 8623923 170635812 8623917
## 6 NA 42.79534 -73.86001 NA 2011-02-11 18:10:08.999 1970-01-01 00:00:00.000 8623923 170635813 8623917
## 7 NA 42.79528 -73.86013 NA 2011-02-11 18:20:58.997 1970-01-01 00:00:00.000 8623923 170635814 8623917
## 9 NA 42.79483 -73.86012 NA 2011-02-11 18:41:30.999 1970-01-01 00:00:00.000 8623923 170635816 8623917
## 15 NA 42.79509 -73.86037 NA 2011-02-11 19:41:10.997 1970-01-01 00:00:00.000 8623923 170635822 8623917
## 16 NA 42.79515 -73.86023 NA 2011-02-11 19:50:58.000 1970-01-01 00:00:00.000 8623923 170635823 8623917
## sensor_type
## 5 GPS
## 6 GPS
## 7 GPS
## 9 GPS
## 15 GPS
## 16 GPSNote: If there are duplicate animal-timestamp records in Movebank, you will get a warning. You can exclude duplicate records on import using removeDuplicatedTimestamps=T. If you are a data manager for a study in Movebank you can also filter them out directly in the study so they are excluded by default in downloads (see https://www.movebank.org/node/27252). Create a data frame from the MoveStack object
fisher.dat <- as(fisher.move, "data.frame")Data cleaning
Delete observations where missing lat or long or a timestamp. There are no missing observations in this data set, but it is still good practice to check.
ind<-complete.cases(fisher.dat[,c("location_lat", "location_long", "timestamp")])
fisher.dat<-fisher.dat[ind==TRUE,]Check for duplicated observations (ones with same lat, long, timestamp, and individual identifier). There are no duplicate observations in this data set, but it is still good practice to check.
ind2<-fisher.dat %>% select(timestamp, location_long, location_lat, local_identifier) %>%
duplicated
sum(ind2) # no duplicates## [1] 0fisher.dat<-fisher.dat[ind2!=TRUE,]Make timestamp a date/time variable
fisher.dat$timestamp<-as.POSIXct(fisher.dat$timestamp, format="%Y-%m-%d %H:%M:%OS", tz="UTC")Look at functions in the move package.
plot(fisher.move)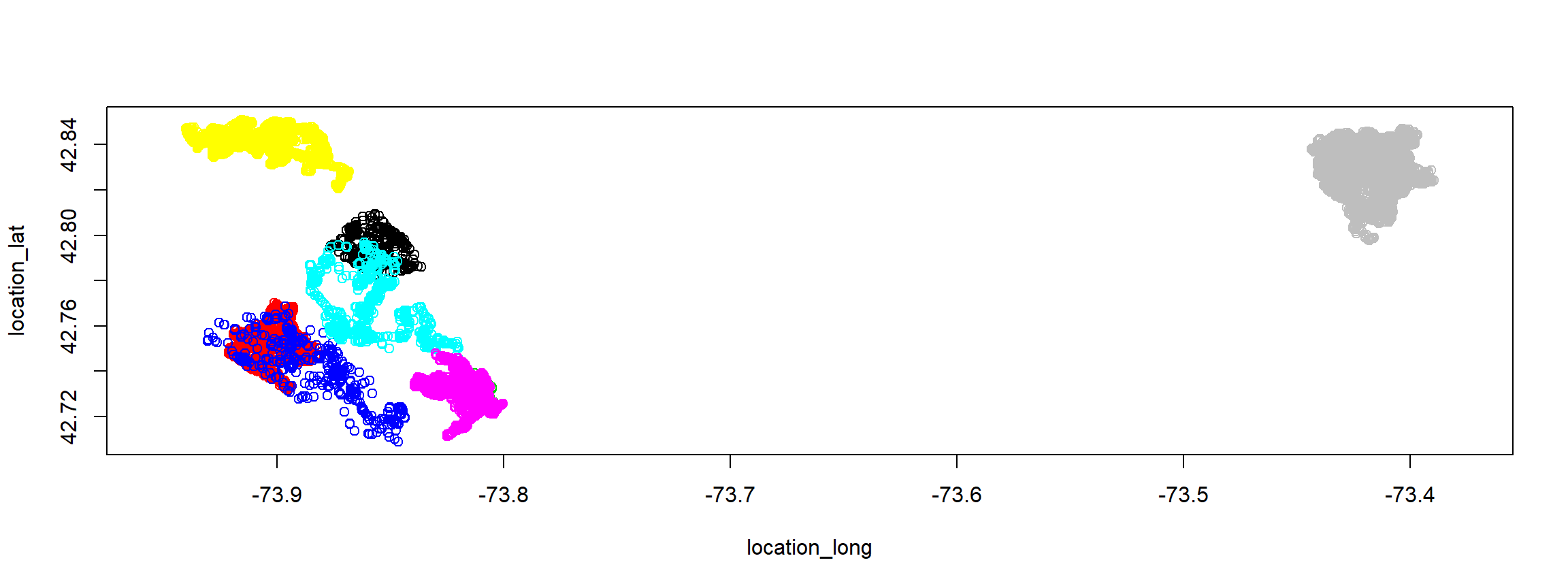
show(fisher.move)## class : MoveStack
## features : 32904
## extent : -73.94032, -73.38947, 42.70898, 42.851 (xmin, xmax, ymin, ymax)
## coord. ref. : +proj=longlat +ellps=WGS84 +datum=WGS84 +towgs84=0,0,0
## variables : 24
## names : sensor_type_id, behavioural_classification, eobs_battery_voltage, eobs_fix_battery_voltage, eobs_horizontal_accuracy_estimate, eobs_key_bin_checksum, eobs_speed_accuracy_estimate, eobs_start_timestamp, eobs_status, eobs_temperature, eobs_type_of_fix, eobs_used_time_to_get_fix, ground_speed, heading, height_above_ellipsoid, ...
## min values : 653, , NA, NA, NA, NA, NA, NA, NA, NA, NA, 3, NA, NA, NA, ...
## max values : 653, traveling, NA, NA, NA, NA, NA, NA, NA, NA, NA, 120, NA, NA, NA, ...
## timestamps : 2009-02-11 12:16:45.000 ... 2011-05-28 09:10:24.999 Time difference of 836 days (start ... end, duration)
## sensors : GPS
## indiv. data : comments, death_comments, earliest_date_born, exact_date_of_birth, individual_id, latest_date_born, local_identifier, nick_name, ring_id, sex, taxon_canonical_name
## min ID Data : "Leroy", adult male, 5.6kg. Fixed temporal schedule GPS tag = 15 minutes., , NA, NA, 8623916, NA, F1, NA, NA, f, NA
## max ID Data : "Zissou", subadult female, 2.35kg. Accelerometer-informed GPS tag, temporal schedule = 2 to 60 minute interval., 7 March 2011 road-killed at 42.752050, -73.820170, NA, NA, 8630581, NA, M5, NA, NA, m, NA
## individuals : F1, F2, F3, M1, M2, M3, M4, M5
## unused rec. : 14443
## license : These data have been published by the Movebank Data Repository with DOI 10.5441/001/1.2tp2j43g. See www.datarepository.movebank.org/handle/10255/mo ...
## citation : LaPoint, S, Gallery P, Wikelski M, Kays R (2013) Animal behavior, cost-based corridor models, and real corridors. Landscape Ecology, v 28 i 8, p 1615–1630. doi:10.1007/s10980-013-9910-0
## study name : Martes pennanti LaPoint New York
## date created: 2018-05-01 17:05:14.051summary(fisher.move)## Object of class MoveStack
## Coordinates:
## min max
## location_long -73.94032 -73.38947
## location_lat 42.70898 42.85100
## Is projected: FALSE
## proj4string :
## [+proj=longlat +ellps=WGS84 +datum=WGS84 +towgs84=0,0,0]
## Number of points: 32904
## Data attributes:
## sensor_type_id behavioural_classification eobs_battery_voltage eobs_fix_battery_voltage eobs_horizontal_accuracy_estimate eobs_key_bin_checksum
## Min. :653 :30468 Mode:logical Mode:logical Mode:logical Mode:logical
## 1st Qu.:653 active : 551 NA's:32904 NA's:32904 NA's:32904 NA's:32904
## Median :653 hunting : 1521
## Mean :653 resting : 63
## 3rd Qu.:653 traveling: 301
## Max. :653
## eobs_speed_accuracy_estimate eobs_start_timestamp eobs_status eobs_temperature eobs_type_of_fix eobs_used_time_to_get_fix ground_speed heading
## Mode:logical Mode:logical Mode:logical Mode:logical Mode:logical Min. : 3.00 Mode:logical Mode:logical
## NA's:32904 NA's:32904 NA's:32904 NA's:32904 NA's:32904 1st Qu.: 7.00 NA's:32904 NA's:32904
## Median : 13.00
## Mean : 24.74
## 3rd Qu.: 37.00
## Max. :120.00
## height_above_ellipsoid location_lat location_long manually_marked_outlier timestamp update_ts deployment_id
## Mode:logical Min. :42.71 Min. :-73.94 Mode:logical Min. :2009-02-11 12:16:45.0 1970-01-01 00:00:00.000:32904 Min. :8623923
## NA's:32904 1st Qu.:42.76 1st Qu.:-73.90 NA's:32904 1st Qu.:2010-03-23 03:57:38.0 1st Qu.:8624627
## Median :42.83 Median :-73.82 Median :2011-02-08 09:33:13.5 Median :8625155
## Mean :42.81 Mean :-73.69 Mean :2010-11-15 07:50:30.5 Mean :8627068
## 3rd Qu.:42.84 3rd Qu.:-73.42 3rd Qu.:2011-04-17 08:12:59.5 3rd Qu.:8630587
## Max. :42.85 Max. :-73.39 Max. :2011-05-28 09:10:25.0 Max. :8630587
## event_id tag_id sensor_type
## Min. :170635812 Min. :8623917 GPS:32904
## 1st Qu.:170652431 1st Qu.:8624568
## Median :170664696 Median :8624887
## Mean :170671625 Mean :8626969
## 3rd Qu.:170696240 3rd Qu.:8630582
## Max. :170705307 Max. :8630582Plots of the data
Note: there are lots of ways to plot data using R. I have included code that illustrates 2 simple plotting methods for a single individual:
- using ggmap (with google maps)
- using leaflet
Both can become cumbersome and slow with too many observations. Lets look at the data from F2. Note: When reading data from Movebank always be sure to refer to the animal-id and individual-local-identifer and not the tag-id or tag-local-identifier in order to exclude pre- and post-deployment records and correctly separate out individual animals when the study includes redeployments.
fisherF2<-fisher.dat %>% filter(local_identifier=="F2")We can plot the data using ggmap and ggplot; ggmap extends ggplot2 by adding maps from e.g. google maps as background Get the map that covers the study area. Zoom out a bit from what calc_zoom suggests. For your own data, you may have to change the zoom.
z<-(calc_zoom(location_long, location_lat, fisherF2))
map <- get_map(location = c(lon = mean(fisherF2$location_long),
lat = mean(fisherF2$location_lat)), zoom = 12,
maptype = "hybrid", source = "google")## Map from URL : http://maps.googleapis.com/maps/api/staticmap?center=42.751612,-73.902532&zoom=12&size=640x640&scale=2&maptype=hybrid&language=en-EN&sensor=falseggmap(map) +
geom_point(data=fisherF2, aes(x=location_long, y=location_lat), size=2.5)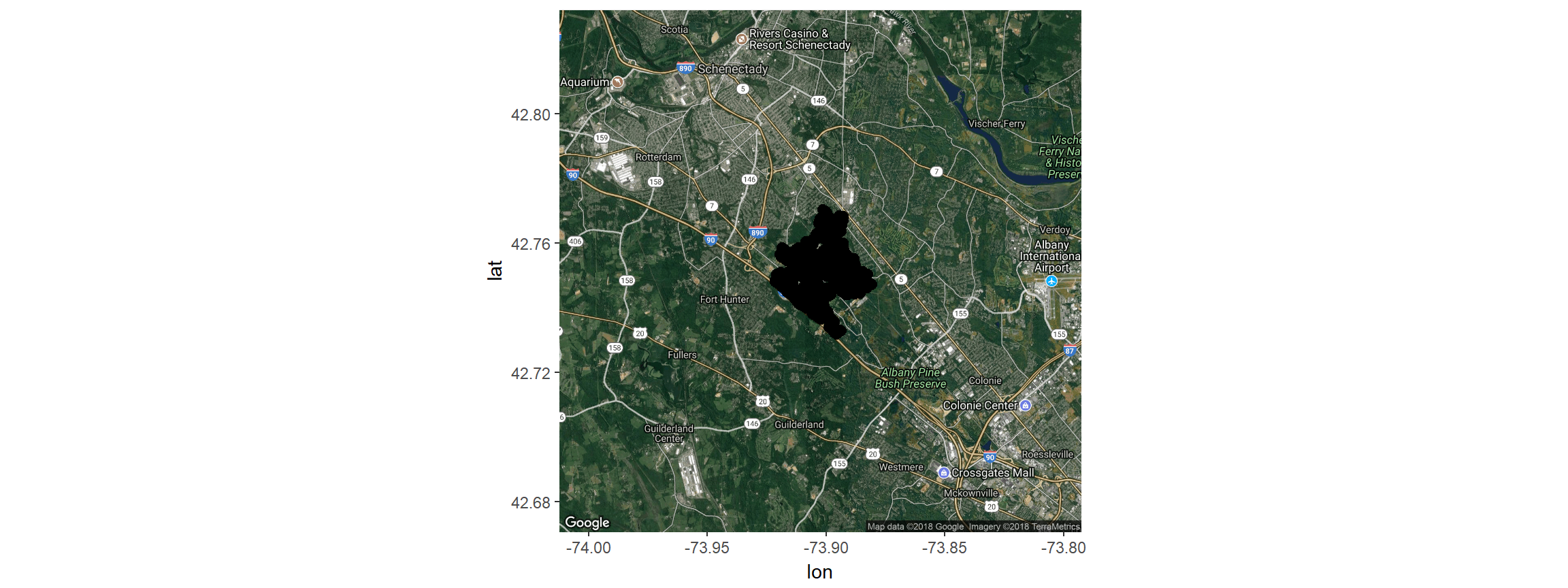
# plot the track on the mapNow, using leaflet
leaflet(fisherF2)%>%addTiles()%>%
addCircles(fisherF2$location_long, fisherF2$location_lat)Using ggplot without a background
Use separate axes for each individual (add scales=“free” to facet_wrap)
ggplot(fisher.dat, aes(x=location_long, y=location_lat))+geom_point()+
facet_wrap(~local_identifier, scales="free")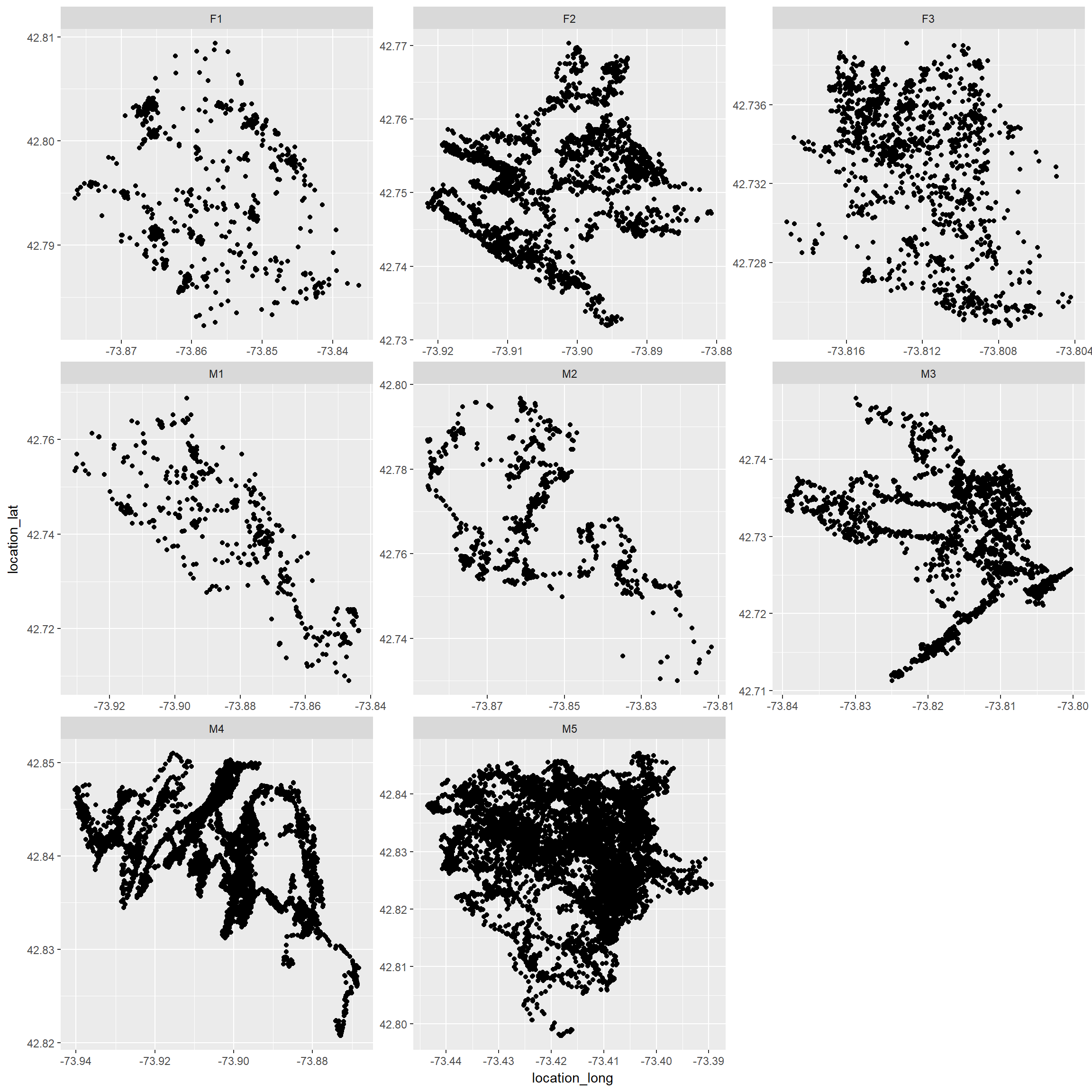
Now, all on 1 plot
ggplot(fisher.dat, aes(x=location_long, y=location_lat, color=as.factor(local_identifier)))+
geom_point() 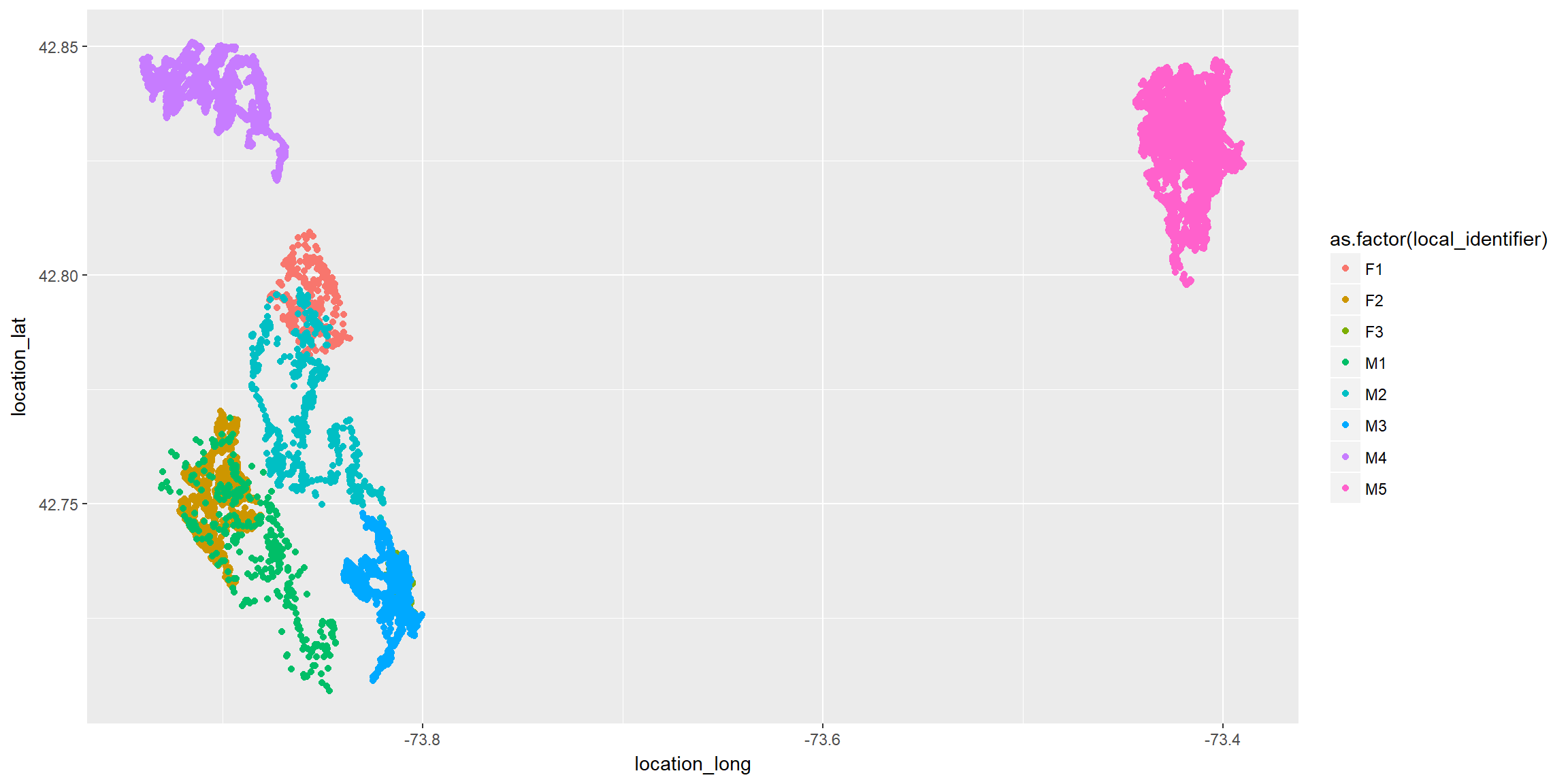
Creating a track in amt
Before we can use the amt package to calculate step lengths, turn angles, and bearings for fisher data, we need to add a class (track) to the data. Then, we can summarize the data by individual, month, etc. First, create a track using utms and the timestamp.
If we have a data set with locations in utms, we could use:
#trk.temp <- make_track(fisher.dat, .x=utm.easting, .y=utm.northing, .t=timestamp, id = individual_local.identifier)
#trk.tempNote: we did not need to explicitly specify x, y and t (but probably good to do so). This would also have worked trk <- make_track(fisher.dat, utm.easting, utm.northing, timestamp, id = local_identifier) We can also use lat, long, which will allow us to determine time of day
trk <- mk_track(fisher.dat, .x=location_long, .y=location_lat, .t=timestamp, id = local_identifier,
crs = CRS("+init=epsg:4326"))## .t found, creating `track_xyt`.# Now it is easy to calculate day/night with either movement track
trk <- trk %>% time_of_day()Now, we can transform back to geographic coordinates
trk <- transform_coords(trk, CRS("+init=epsg:32618"))If we create trk.temp as above, we can make sure that we got back the same coordinates by comparing to the original track
#mean(trk$x_ - trk.temp$x_) # look good, there is virtually no difference
#mean(trk$y_ - trk.temp$y_) # same for ySave the class here (and apply it later after adding columns to the object)
trk.class<-class(trk)Movement Characteristics
- dir_abs will calculate absolute angles for steps
- dir_rel will calculate turning angles (relative angles)
- step_lengths will calculate distances between points
- nsd = Net Squared Displacement (distance from first point)
Arguments direction_abs:
- full_circle will calculate between 0 and 360 (rather than -180 and 180)
- zero gives the direction = 0 for absolute angle
Note: we have to calculate these characteristics separately for each individual (to avoid calculating a distance between say the last observation of the first individual and the first observation of the second individual).
To do this, we could loop through individuals, calculate these characteristics for each individual, then rbind the data back together. Or, use nested data frames and the map function in the purrr library to do this with very little code.
To see how nesting works, we can create a nested object by individual
nesttrk<-trk%>%nest(-id)
nesttrk## # A tibble: 8 x 2
## id data
## <fct> <list>
## 1 M1 <tibble [919 x 4]>
## 2 M4 <tibble [8,958 x 4]>
## 3 F2 <tibble [3,004 x 4]>
## 4 M3 <tibble [2,436 x 4]>
## 5 F3 <tibble [1,501 x 4]>
## 6 M2 <tibble [1,638 x 4]>
## 7 F1 <tibble [1,349 x 4]>
## 8 M5 <tibble [13,099 x 4]>Each row contains data from an individual. For example, we can access data from the first individual using:
nesttrk$data[[1]]## # A tibble: 919 x 4
## x_ y_ t_ tod_
## * <dbl> <dbl> <dttm> <chr>
## 1 590130. 4732942. 2009-02-11 12:16:45.000 day
## 2 590136. 4732940. 2009-02-11 12:31:38.000 day
## 3 590138. 4732935. 2009-02-11 12:45:48.997 day
## 4 590144. 4732947. 2009-02-11 13:00:16.002 day
## 5 590137. 4732940. 2009-02-11 13:15:19.000 day
## 6 590126. 4732936. 2009-02-11 13:30:13.000 day
## 7 590139. 4732941. 2009-02-11 13:45:37.000 day
## 8 590139. 4732923. 2009-02-11 14:00:35.000 day
## 9 590137. 4732934. 2009-02-11 14:15:49.000 day
## 10 590136. 4732935. 2009-02-11 14:30:49.000 day
## # ... with 909 more rowsWe could calculate movement characteristics by individual using:
temp<-direction_rel(nesttrk$data[[1]])
head(temp)## [1] NA -55.96892 130.93489 159.69985 -24.84488 179.16947or:
temp<-trk %>% filter(id=="M1") %>% direction_rel
head(temp)## [1] NA -55.96892 130.93489 159.69985 -24.84488 179.16947Or, we can add a columns to each nested column of data using purrr::map
trk<-trk %>% nest(-id) %>%
mutate(dir_abs = map(data, direction_abs,full_circle=TRUE, zero="N"),
dir_rel = map(data, direction_rel),
sl = map(data, step_lengths),
nsd_=map(data, nsd))%>%unnest()Now, calculate month, year, hour, week of each observation and append these to the dataset Unlike the movement charactersitics, these calculations can be done all at once, since they do not utilize successive observations (like step lengths and turn angles do).
trk<-trk%>%
mutate(
week=week(t_),
month = month(t_, label=TRUE),
year=year(t_),
hour = hour(t_)
)Now, we need to again tell R that this is a track (rather than just a data frame)
class(trk)## [1] "tbl_df" "tbl" "data.frame"class(trk)<-trk.classLets take a look at what we created
trk## # A tibble: 32,904 x 13
## id dir_abs dir_rel sl nsd_ x_ y_ t_ tod_ week month year hour
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dttm> <chr> <dbl> <ord> <dbl> <int>
## 1 M1 259. NA 6.38 0 590130. 4732942. 2009-02-11 12:16:45.000 day 6 Feb 2009 12
## 2 M1 203. -56.0 5.61 40.7 590136. 4732940. 2009-02-11 12:31:38.000 day 6 Feb 2009 12
## 3 M1 334. 131. 12.7 112. 590138. 4732935. 2009-02-11 12:45:48.997 day 6 Feb 2009 12
## 4 M1 134. 160. 9.65 222. 590144. 4732947. 2009-02-11 13:00:16.002 day 6 Feb 2009 13
## 5 M1 109. -24.8 11.7 52.2 590137. 4732940. 2009-02-11 13:15:19.000 day 6 Feb 2009 13
## 6 M1 288. 179. 13.9 46.1 590126. 4732936. 2009-02-11 13:30:13.000 day 6 Feb 2009 13
## 7 M1 180. -109. 17.3 85.5 590139. 4732941. 2009-02-11 13:45:37.000 day 6 Feb 2009 13
## 8 M1 13.0 -167. 10.5 419. 590139. 4732923. 2009-02-11 14:00:35.000 day 6 Feb 2009 14
## 9 M1 34.7 21.7 1.57 111. 590137. 4732934. 2009-02-11 14:15:49.000 day 6 Feb 2009 14
## 10 M1 317. -77.8 7.51 80.7 590136. 4732935. 2009-02-11 14:30:49.000 day 6 Feb 2009 14
## # ... with 32,894 more rowsSome plots of movement characteristics
Absolute angles (for each movement) relative to North
We could use a rose diagram (below) to depict the distribution of angles.
ggplot(trk, aes(x = dir_abs, y=..density..)) + geom_histogram(breaks = seq(0,360, by=20))+
coord_polar(start = 0) + theme_minimal() +
scale_fill_brewer() + ylab("Density") + ggtitle("Angles Direct") +
scale_x_continuous("", limits = c(0, 360), breaks = seq(0, 360, by=20),
labels = seq(0, 360, by=20))+
facet_wrap(~id)## Warning: Removed 8 rows containing non-finite values (stat_bin).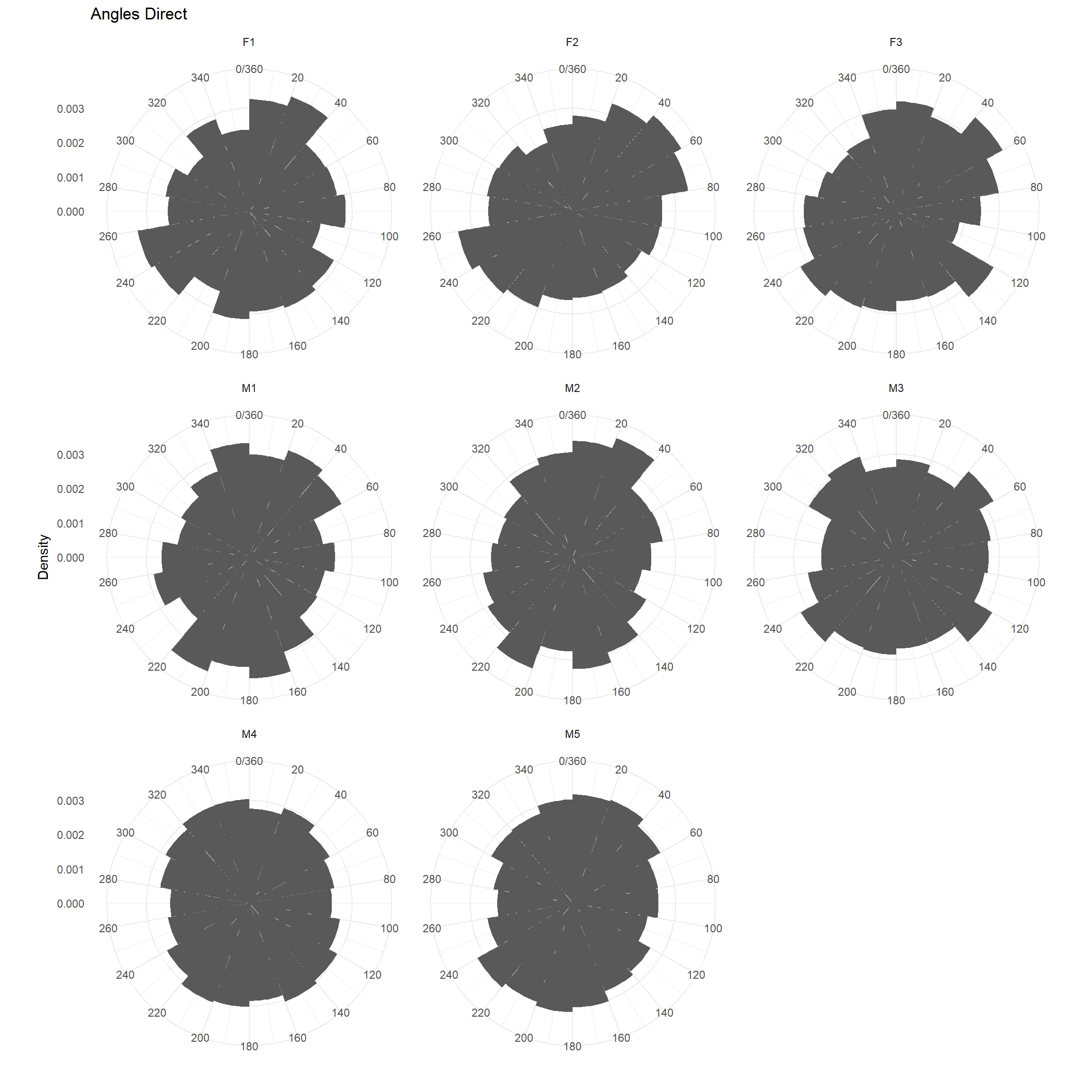
Turning angles
Note: a 0 indicates the animal continued to move in a straight line, a 180 indicates the animal turned around (but note, resting + measurement error often can make it look like the animal turned around).
ggplot(trk, aes(x = dir_rel, y=..density..)) + geom_histogram(breaks = seq(-180,180, by=20))+
coord_polar(start = 0) + theme_minimal() +
scale_fill_brewer() + ylab("Density") + ggtitle("Angles Direct") +
scale_x_continuous("", limits = c(-180, 180), breaks = seq(-180, 180, by=20),
labels = seq(-180, 180, by=20))+
facet_wrap(~id)## Warning: Removed 16 rows containing non-finite values (stat_bin).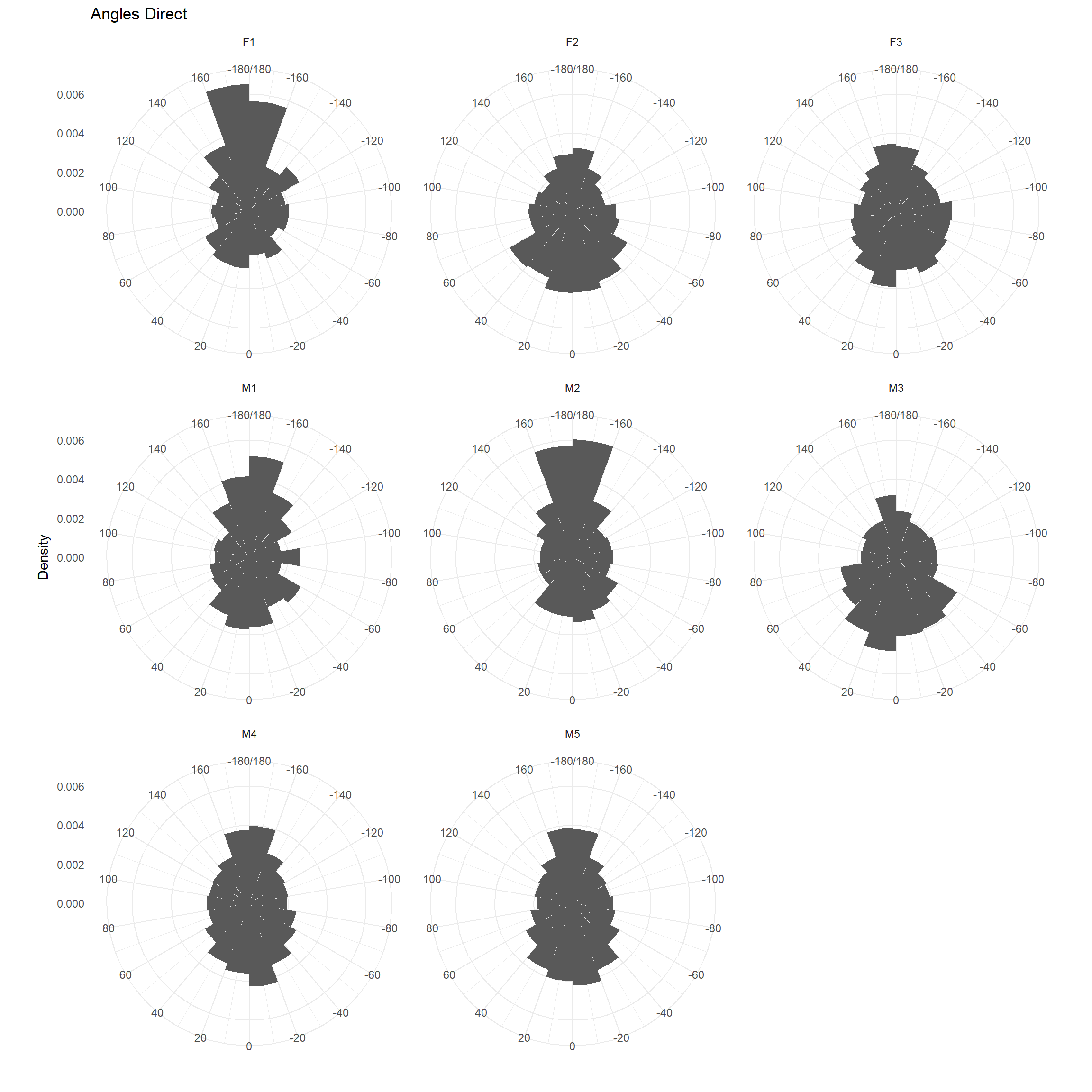
Turning angles as histograms
ggplot(trk, aes(x = dir_rel)) + geom_histogram(breaks = seq(-180,180, by=20))+
theme_minimal() +
scale_fill_brewer() + ylab("Count") + ggtitle("Angles Relative") +
scale_x_continuous("", limits = c(-180, 180), breaks = seq(-180, 180, by=20),
labels = seq(-180, 180, by=20))+facet_wrap(~id, scales="free")## Warning: Removed 16 rows containing non-finite values (stat_bin).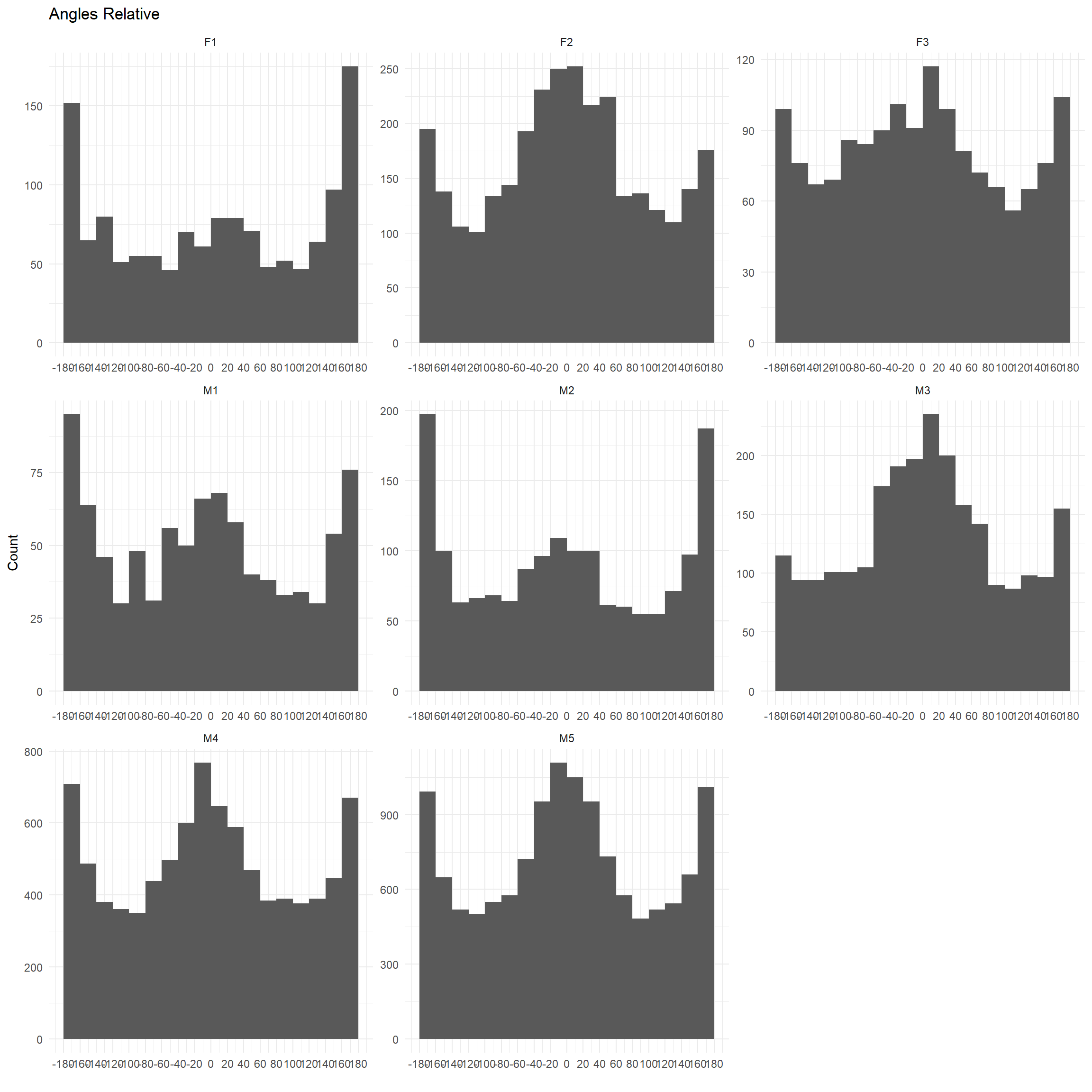
Net-squared displacement over time for each individual
ggplot(trk, aes(x = t_, y=nsd_)) + geom_point()+
facet_wrap(~id, scales="free")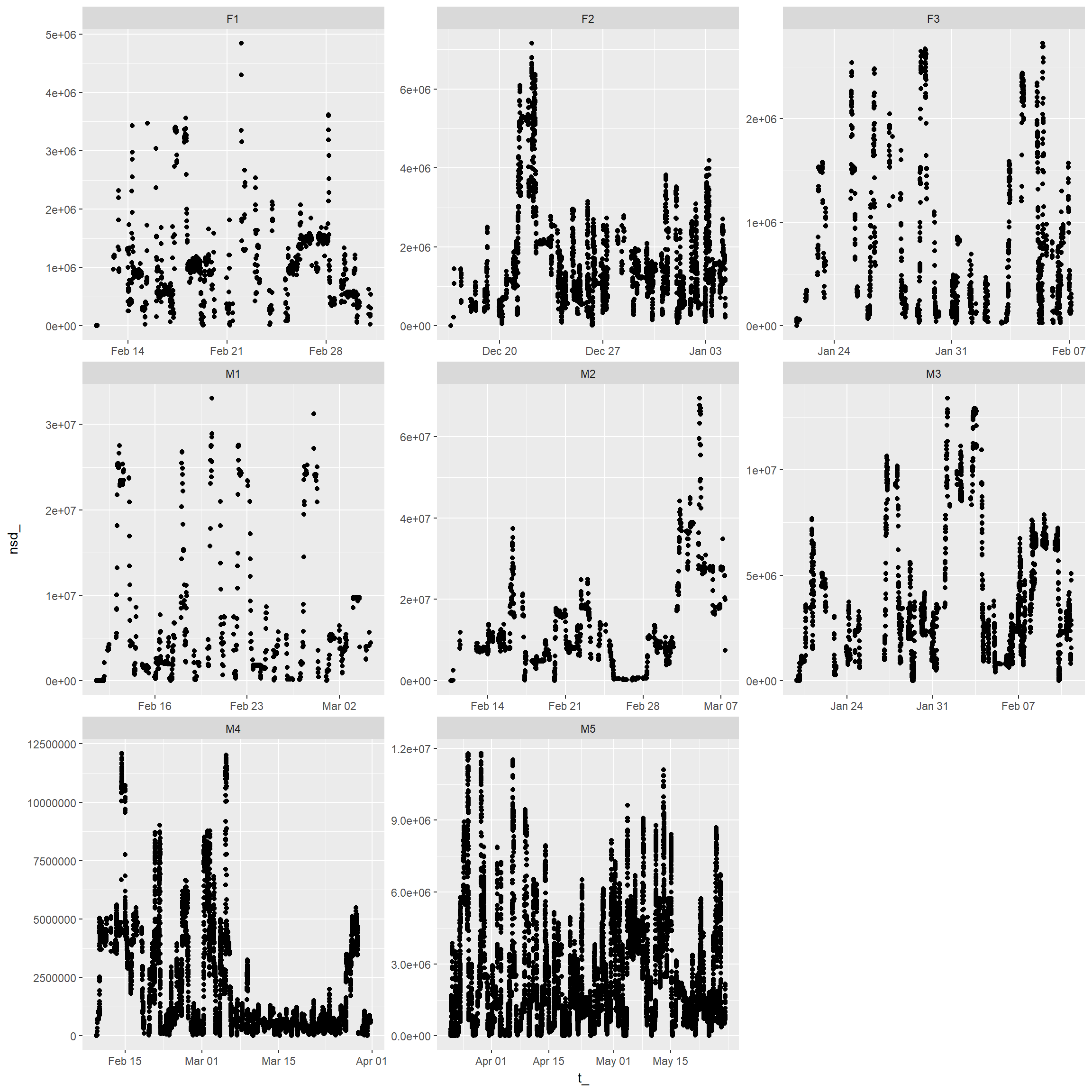
Explore movement characteristics by (day/night, hour, month)
step length distribution by day/night
ggplot(trk, aes(x = tod_, y = log(sl))) +
geom_boxplot()+geom_smooth()+facet_wrap(~id)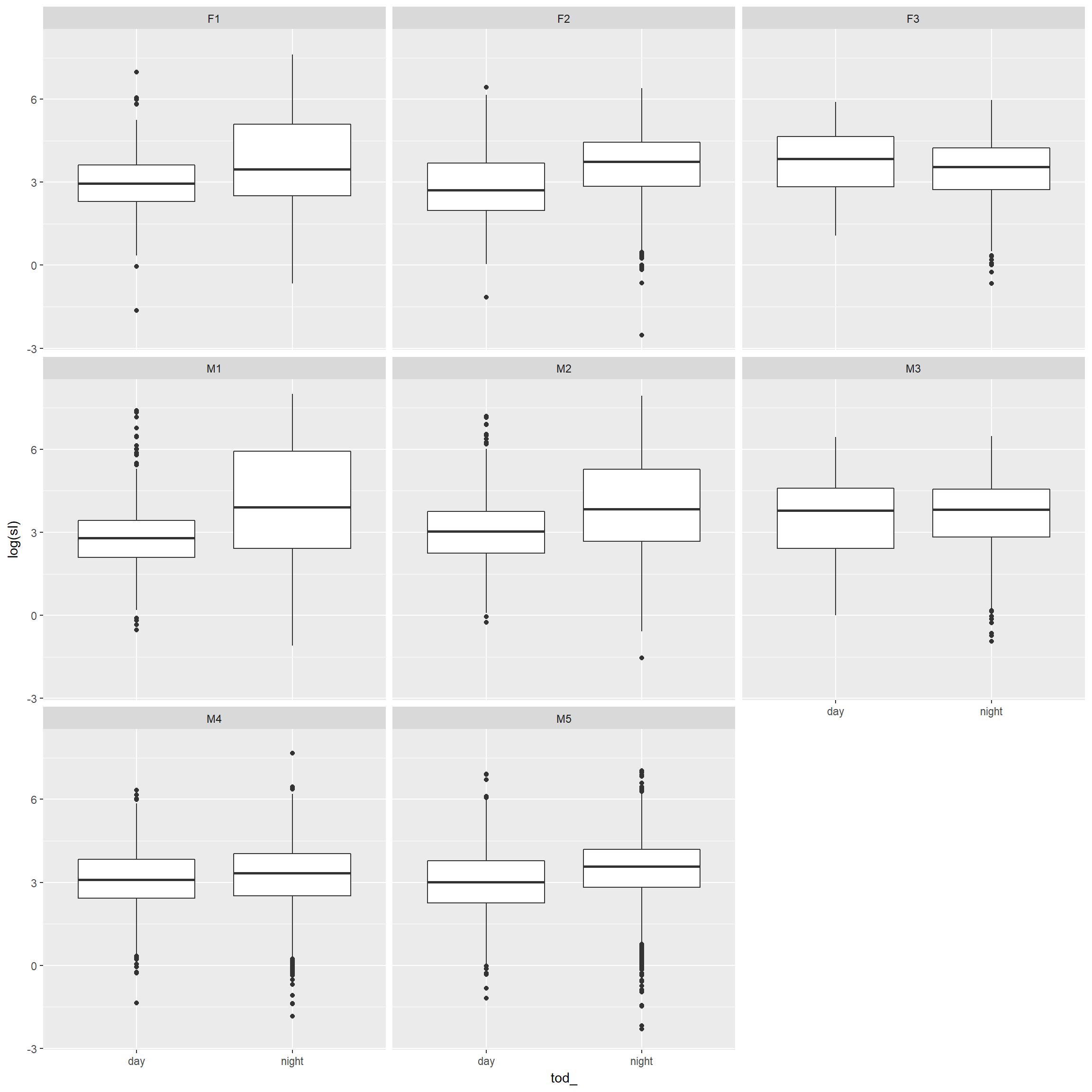
Space use (MCP or KDE) by week, month, and year
Note: this code will only work for your critters if you have multiple observations for each combination of (month, year). If you don’t have many observations, you could try: nest(-id, -year) and unnest(id,year)
mcps.week<-trk %>% nest(-id,-year, -month, -week) %>%
mutate(mcparea = map(data, ~hr_mcp(., levels = c(0.95)) %>% hr_area)) %>%
select(id, year, month, week, mcparea) %>% unnest()ggplot(mcps.week, aes(x = week, y = area, colour=as.factor(year))) + geom_point()+
geom_smooth()+ facet_wrap(~id, scales="free")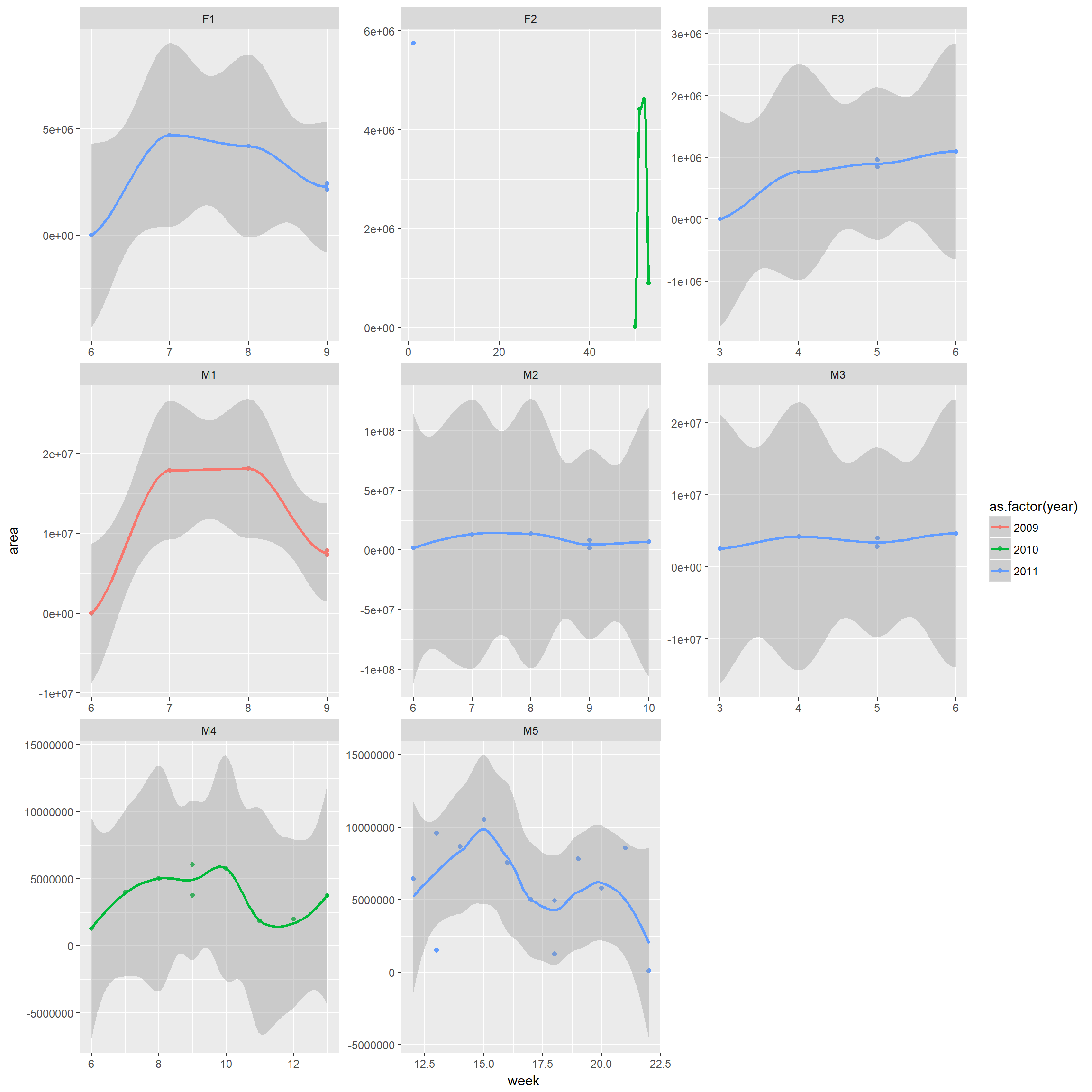
Same for KDE
kde.week<-trk %>% nest(-id,-year, -month, -week) %>%
mutate(kdearea = map(data, ~hr_kde(., levels=c(0.95)) %>% hr_area)) %>%
select(id, year, month, week, kdearea) %>% unnest()ggplot(kde.week, aes(x = week, y = kdearea, colour=as.factor(year))) + geom_point()+
geom_smooth()+ facet_wrap(~id, scales="free")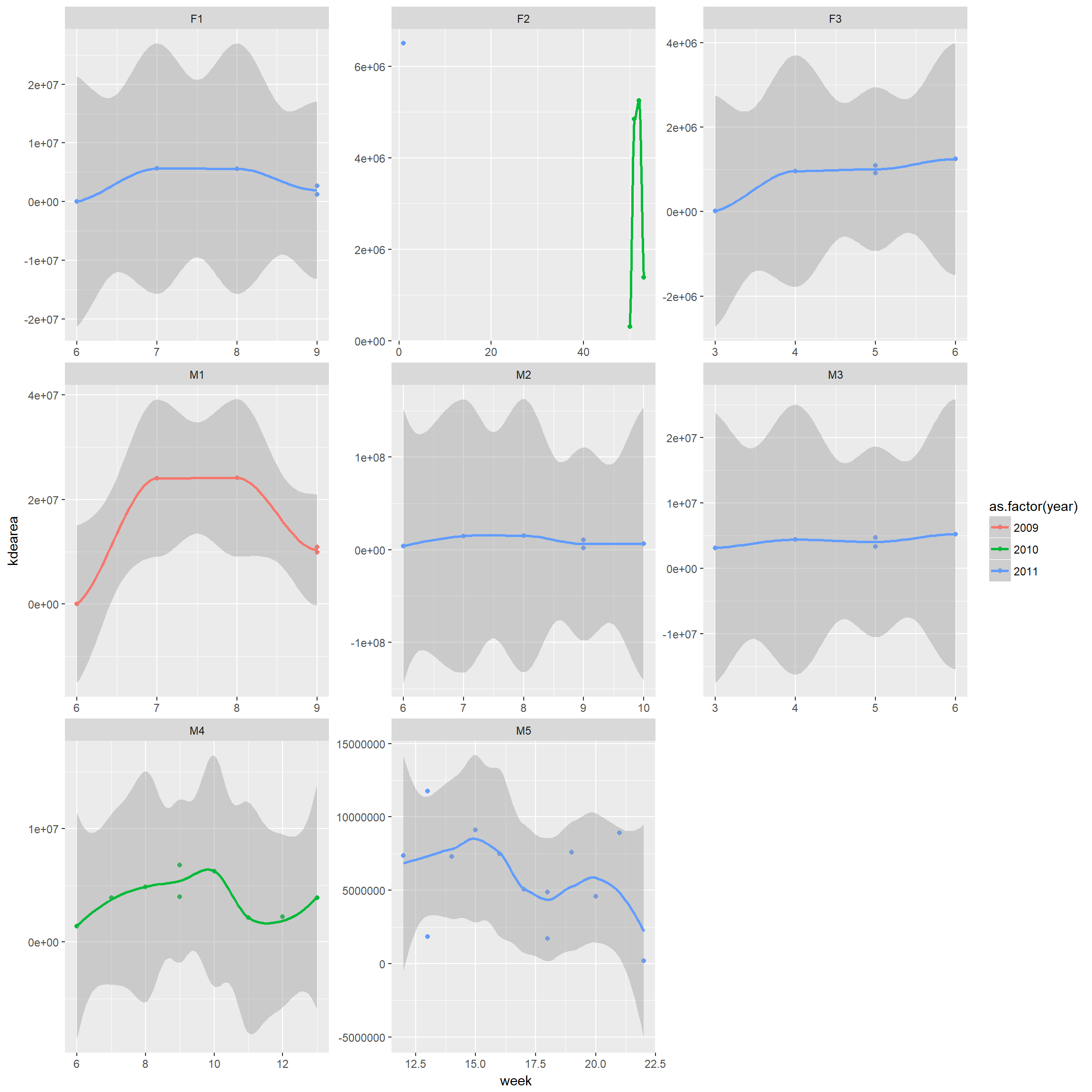
RSF prep
Generate random points within MCPs for a single individual using amt functions. Notes:
- It is common to generate points randomly, but other options are possible.
- In particular, it can beneficial to generate a systematically placed sample
- Samples can also be generated using the spsample function in the sp library or using a GIS (note: amt uses the spsample function within its function random_points)
- Other home range polygons could be used (e.g., kernel density, local convex hull etc.)
Random points: illustrate for 1 individual
trk %>% filter(id=="F1") %>%
random_points(.,factor = 20) %>% plot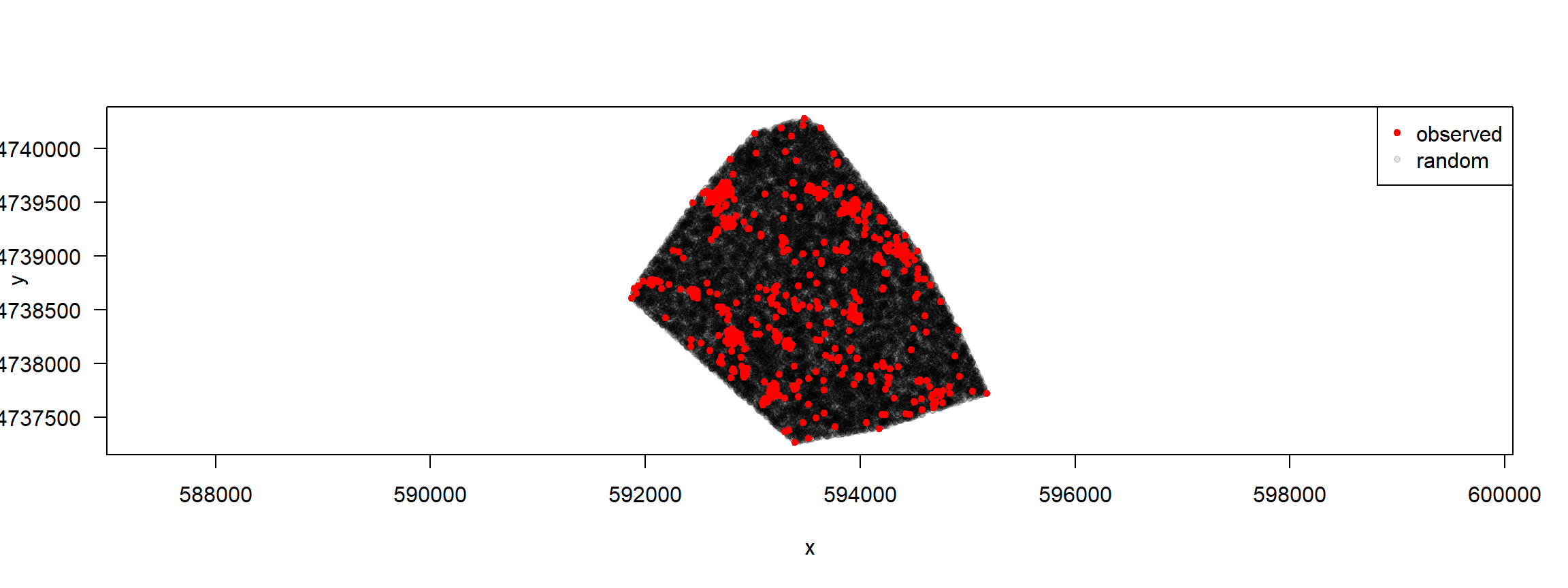
Illustrate systematic points (to do this, we need to create the mcp first)
trk%>%filter(id=="F1") %>%
random_points(., factor = 20, type="regular") %>%
plot() 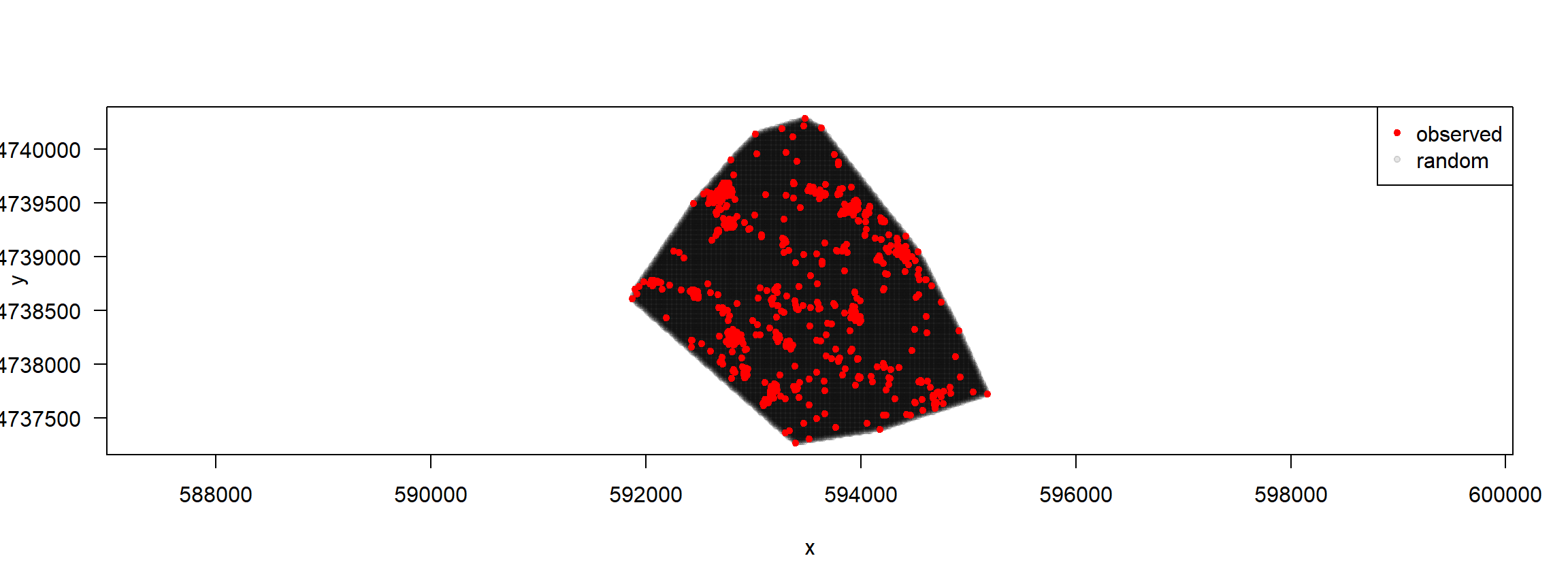
Now, lets generate points for all individuals. We can do this efficiently by making use of pipes (%>%),nested data frames, and then by adding a new column – a list-column – to trks
avail.pts <- trk %>% nest(-id) %>%
mutate(rnd_pts = map(data, ~ random_points(., factor = 20, type="regular"))) %>%
select(id, rnd_pts) %>% # you dont want to have the original point twice, hence drop data
unnest()Or, we could do this using a loop (commented out, below)
#avail.pts<-NULL
#uid<-unique(trk$id) # individual identifiers
#luid<-length(uid) # number of unique individuals
#for(i in 1:luid){
# random_points will generate random points within mcp
# Add on the individual id and combine all data
# temp<-cbind(id=uid[i],trk%>%filter(id==uid[i])%>%random_points)
# avail.pts<-rbind(avail.pts, temp)
#}
#avail.pts<-as_tibble(avail.pts)
#avail.ptsWrite out data for further annotating
Need to rename variables so everything is in the format Movebank requires for annotation of generic time-location records (see https://www.movebank.org/node/6608#envdata_generic_request). This means, we need the following variables:
- location-lat (perhaps with addition of Easting/Northing in UTMs)
- location-long (perhaps with addition of Easting/Northing in UTMs)
- timestamp (in Movebank format)
Need to project to lat/long, while also keeping lat/long. Then rename variables and write out the data sets.
avail <- SpatialPointsDataFrame(avail.pts[,c("x_","y_")], avail.pts,
proj4string=CRS("+proj=utm +zone=18N +datum=WGS84"))
avail.df <- data.frame(spTransform(avail, CRS("+proj=longlat +datum=WGS84")))[,1:6]
names(avail.df)<-c("idr", "case_", "utm.easting", "utm.northing", "location-long", "location-lat")Check to make sure everything looks right
test<-subset(avail.df, case_==TRUE)
test %>% select('location-lat', 'location-long', utm.easting, utm.northing) %>%
summarise_all(mean)## location-lat location-long utm.easting utm.northing
## 1 42.80687 -73.69359 606812.2 4740221fisher.dat %>% summarize(meanloc.lat=mean(location_lat),
meanloc.long=mean(location_long))## meanloc.lat meanloc.long
## 1 42.80687 -73.69359Add a timestamp to annotate these data with environmental covariates in Movebank using Env-DATA (https://www.movebank.org/node/6607). Here we just use the first timestamp, however meaningful timestamps are needed if annotating variables that vary in time.
avail.df$timestamp<-fisher.dat$timestamp[1]These points then need to be annotated prior to fitting rsfs. Let’s write out 2 files:
- FisherRSF2018.csv will contain all points and identifying information.
- FisherRSFannotate.csv will contain only the columns used to create the annotation.
The latter file will take up less space, making it easier to annotate (and also possible to upload to github)
write.csv(avail.df, file="data/FisherRSF2018.csv", row.names = FALSE)
avail.df<-avail.df %>% select("timestamp", "location-long", "location-lat")
write.csv(avail.df, file="data/FisherRSFannotate.csv", row.names = FALSE)SSF prep
SSFs assume that data have been collected at regular time intervals. We can use the track_resample function to regularize the trajectory so that all points are located within some tolerence of each other in time. To figure out a meaningful tolerance range, we should calculate time differences between locations & look at as a function of individual.
(timestats<-trk %>% nest(-id) %>% mutate(sr = map(data, summarize_sampling_rate)) %>%
select(id, sr) %>% unnest)## # A tibble: 8 x 10
## id min q1 median mean q3 max sd n unit
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <chr>
## 1 M1 13.3 14.7 15.0 32.7 15.5 977. 104. 918 min
## 2 M4 0.1000 1.93 2.03 8.04 2.57 1209. 44.0 8957 min
## 3 F2 0.400 1.97 2.03 8.93 3.98 1080. 48.1 3003 min
## 4 M3 0.417 1.97 2.17 13.3 5.42 2889. 90.2 2435 min
## 5 F3 0.417 1.97 2.07 15.7 4.08 2167. 104. 1500 min
## 6 M2 1.35 9.78 10.0 21.7 10.3 2111. 87.2 1637 min
## 7 F1 3.92 9.80 10.0 20.7 10.3 1650. 95.2 1348 min
## 8 M5 0.250 1.93 2.02 7.41 2.37 976. 25.0 13098 minTime intervals range from every 2 to 15 minutes on average, depending on the individual. Lets add on the time difference to each obs.
trk<-trk %>% group_by(id) %>% mutate(dt_ = t_ - lag(t_, default = NA))Let’s illustrate track regularization with ID = F2. Let’s go from every 2 minutes to every 10.
tempF2<-trk %>% filter(id=="F2") %>% track_resample(rate=minutes(10), tolerance=minutes(2))
tempF2 %>% select(id, x_, y_, t_, burst_)## # A tibble: 1,010 x 5
## # Groups: id [1]
## id x_ y_ t_ burst_
## * <fct> <dbl> <dbl> <dttm> <dbl>
## 1 F2 590028. 4733219. 2010-12-16 16:45:30.999 1
## 2 F2 590486. 4733344. 2010-12-16 21:21:13.999 2
## 3 F2 591220. 4733065. 2010-12-16 21:32:34.000 2
## 4 F2 591215. 4733037. 2010-12-17 08:57:00.999 3
## 5 F2 591172. 4733057. 2010-12-17 09:05:14.000 3
## 6 F2 591075. 4733081. 2010-12-17 09:16:29.000 3
## 7 F2 591024. 4733025. 2010-12-17 09:24:53.000 3
## 8 F2 590793. 4732994. 2010-12-17 09:37:24.000 4
## 9 F2 590838. 4733077. 2010-12-18 01:08:42.997 5
## 10 F2 590658. 4733176. 2010-12-18 01:24:34.000 6
## # ... with 1,000 more rowsNow loop over individuals and do the following:
- Regularize trajectories using an appropriate time window (see e.g., below)
- calculate new dt values
- Create bursts using individual-specific time intervals
- Generate random steps within each burst
The random steps are generated using the following approach:
- Fit a gamma distribution to step lenghts
- Fit a von mises distribution to turn angles
- Use these distribution to draw new turns and step lengths, form new simulated steps and generate random x,y values.
ssfdat<-NULL
temptrk<-with(trk, track(x=x_, y=y_, t=t_, id=id))
uid<-unique(trk$id) # individual identifiers
luid<-length(uid) # number of unique individuals
for(i in 1:luid){
# Subset individuals & regularize track
temp<-temptrk%>% filter(id==uid[i]) %>%
track_resample(rate=minutes(round(timestats$median[i])),
tolerance=minutes(max(10,round(timestats$median[i]/5))))
# Get rid of any bursts without at least 2 points
temp<-filter_min_n_burst(temp, 2)
# burst steps
stepstemp<-steps_by_burst(temp)
# create random steps using fitted gamma and von mises distributions and append
rnd_stps <- stepstemp %>% random_steps(n = 15)
# append id
rnd_stps<-rnd_stps%>%mutate(id=uid[i])
# append new data to data from other individuals
ssfdat<-rbind(rnd_stps, ssfdat)
}
ssfdat<-as_tibble(ssfdat)
ssfdat## # A tibble: 470,000 x 13
## burst_ step_id_ case_ x1_ y1_ x2_ y2_ t1_ t2_ dt_ sl_ ta_ id
## <dbl> <int> <lgl> <dbl> <dbl> <dbl> <dbl> <dttm> <dttm> <time> <dbl> <dbl> <fct>
## 1 1 1 TRUE 630498. 4742879. 630569. 4742823. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 90.4 -62.6 M5
## 2 1 1 FALSE 630498. 4742879. 630531. 4742824. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 33.3 -113. M5
## 3 1 1 FALSE 630498. 4742879. 630509. 4742929. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 107. -130. M5
## 4 1 1 FALSE 630498. 4742879. 630500. 4742826. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 3.90 -125. M5
## 5 1 1 FALSE 630498. 4742879. 630506. 4742823. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 7.73 50.2 M5
## 6 1 1 FALSE 630498. 4742879. 630473. 4742855. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 40.4 77.6 M5
## 7 1 1 FALSE 630498. 4742879. 630526. 4742862. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 48.2 -150. M5
## 8 1 1 FALSE 630498. 4742879. 630574. 4742643. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 195. 143. M5
## 9 1 1 FALSE 630498. 4742879. 630534. 4742723. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 107. 80.5 M5
## 10 1 1 FALSE 630498. 4742879. 630495. 4742822. 2011-03-22 00:02:12.997 2011-03-22 00:04:10.997 118 3.41 28.8 M5
## # ... with 469,990 more rowsNow, lets plot the data for random and matched points
ggplot(ssfdat, aes(x2_, y2_, color=case_))+geom_point()+facet_wrap(~id, scales="free")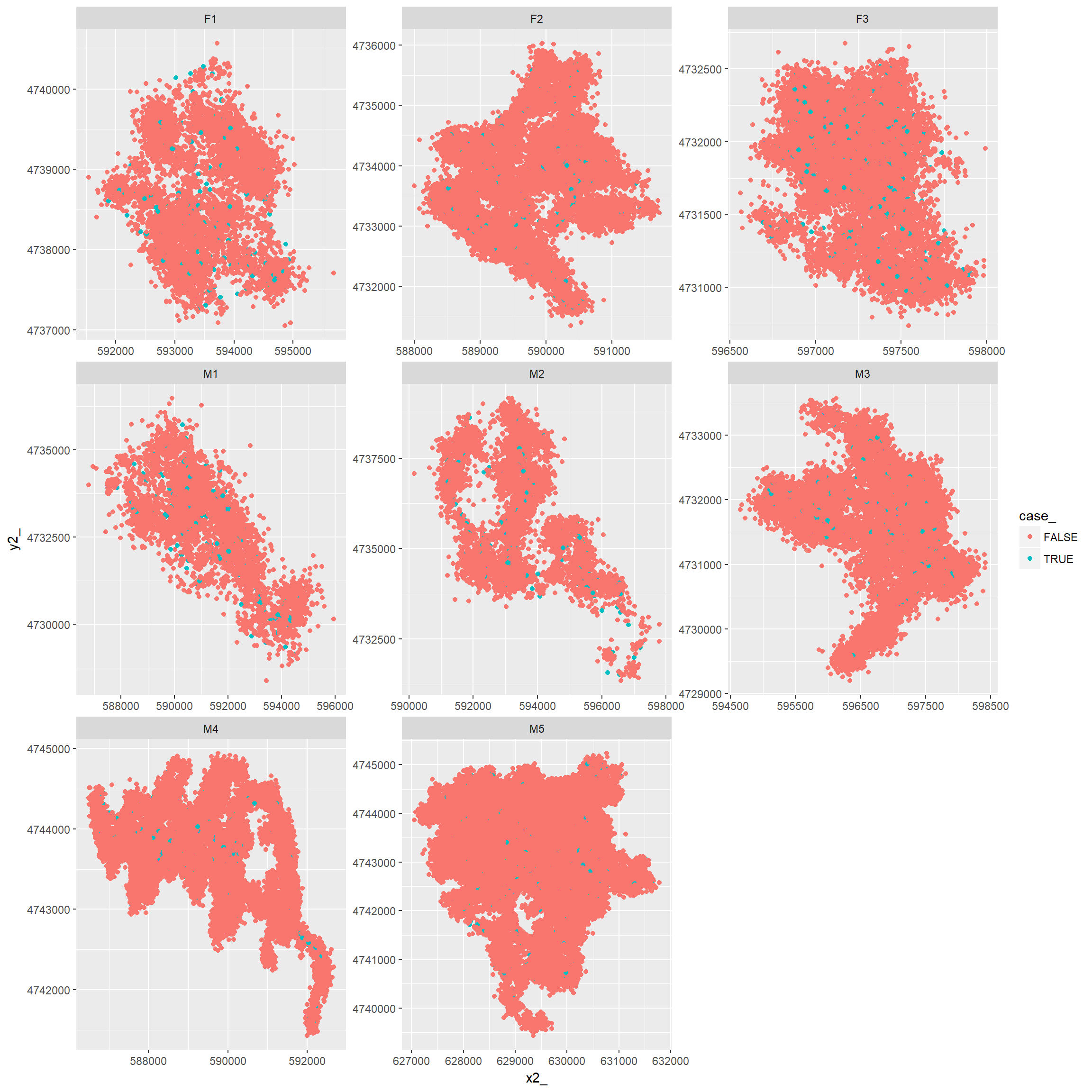
Relabel as utms
ssfdat$utm.easting<-ssfdat$x2_
ssfdat$utm.northing<-ssfdat$y2_Write out data for further annotating
Need to rename variables so everything is in the format Movebank requires for annotation of generic time-location records (see https://www.movebank.org/node/6608#envdata_generic_request). This means, we need the following variables:
- location-lat (perhaps with addition of Easting/Northing in UTMs)
- location-long (perhaps with addition of Easting/Northing in UTMs)
- timestamp (in Movebank format)
Need to project to lat/long, while also keeping lat/long. Then rename variables and write out the data sets. With the SSFs, we have the extra complication of having a time and location at both the start and end of the step.
For the time being, we will assume we want to annotate variables at the end of the step but use the starting point of the step as the timestamp.
You could also calculate the midpoint of the timestep like this: data$timestamp.midpoint <- begintime + (endtime-begintime)/2
ssfdat2 <- SpatialPointsDataFrame(ssfdat[,c("x2_","y2_")], ssfdat,
proj4string=CRS("+proj=utm +zone=18N +datum=WGS84"))
ssf.df <- data.frame(spTransform(ssfdat2, CRS("+proj=longlat +datum=WGS84")))
names(ssf.df)[c(13,16,17)] <-c("individual.local.identifier", "location-long", "location-lat")
ssf.df$timestamp<-ssf.df$t1_
ssf.df %>% select('location-lat', utm.easting, x1_, x2_, y1_, y2_, 'location-long', utm.northing) %>% head## location-lat utm.easting x1_ x2_ y1_ y2_ location-long utm.northing
## 1 42.82684 630568.9 630497.9 630568.9 4742879 4742823 -73.40259 4742823
## 2 42.82685 630531.2 630497.9 630531.2 4742879 4742824 -73.40305 4742824
## 3 42.82780 630509.4 630497.9 630509.4 4742879 4742929 -73.40329 4742929
## 4 42.82688 630500.4 630497.9 630500.4 4742879 4742826 -73.40342 4742826
## 5 42.82685 630505.6 630497.9 630505.6 4742879 4742823 -73.40336 4742823
## 6 42.82714 630472.7 630497.9 630472.7 4742879 4742855 -73.40375 4742855These points then need to be annotated prior to fitting ssfs. Let’s write out 2 files:
- FisherSSF2018.csv will contain all points and identifying information.
- FisherSSFannotate.csv will contain only the columns used to create the annotation.
The latter file will take up less space, making it easier to annotate (and also possible to upload to github)
write.csv(ssf.df, file="data/AllStepsFisher2018.csv", row.names=FALSE)
ssf.df<-ssf.df %>% select("timestamp", "location-long", "location-lat")
write.csv(ssf.df, file="data/FisherSSFannotate.csv", row.names = FALSE)Using nested data frames
This works if all animals are sampled at a constant sampling rate. Again, you need to create a new column (using mutate) where you save the random steps to Not all animals have a 15 min sampling rate, so we might drop the first animal that has a 15 minute sampling rate.
#trk %>% nest(-id) %>% mutate(sr = map(.$data, summarize_sampling_rate)) %>%
# select(id, sr) %>% unnest()
# Then, we could avoid a loop with:
#ssfdat <- trk %>% filter(id != "M1") %>% nest(-id) %>%
# mutate(ssf = map(data, function(d) {
# d %>%
# track_resample(rate = minutes(10), tolerance = minutes(2)) %>%
# filter_min_n_burst(min_n = 3) %>%
# steps_by_burst() %>% random_steps()
# })) %>% select(id, ssf) %>% unnest()








